Abstract
Ewing tumors are a clinically heterogeneous group of childhood sarcomas that represent a paradigm for understanding solid tumor biology, as they are the first group of sarcomas for which a chromosome translocation has been characterized at the molecular level. However, the biologic organization of the tumor, especially the processes that govern proliferation, differentiation, and metastasis of primitive tumor stem cells is poorly understood. Therefore, to develop a biologically relevant in vivo model, five different Ewing tumor cell lines and primary tumor cells from three patients were transplanted into immune-deficient mice via intravenous injection. NOD/ scid mice that carry a complex immune deficiency and thus nearly completely lack the ability to reject human cells were used as recipients. Overall, 26 of 52 mice (50%) transplanted with VH-64, WE-68, CADO-ES1, TC-71, and RM-82 cells and 4 of 10 mice (40%) transplanted with primary tumor cells engrafted. Moreover, primary cells that did not grow in vitro proliferated in mice. The pattern of metastasis was similar to that in patients with frequent metastases in lungs (62%), bone marrow (30%), and bone (23%). Using limiting dilution experiments, the frequency of the engraftment unit was estimated at 1 Ewing tumor-initiating cell in 3 × 105 VH-64 cells. These data demonstrate that we have been able to establish an in vivo model that recapitulates many aspects of growth and progression of human Ewing tumors. For the first time, this model provides the opportunity to identify and characterize primitive in vivo clonogenic solid tumor stem cells. This model will, therefore, be instrumental in studying many aspects of tumor cell biology, including organ-selective metastasis and tumor angiogenesis.
Similar content being viewed by others
Main
The family of Ewing tumors is a clinically heterogeneous group of tumors including Ewing's sarcoma of the bone, Askin tumors of the thoracic wall, and PNET affecting children, adolescents, and young adults. Despite this clinical heterogeneity, Ewing tumors represent a paradigm for understanding the biology of human solid tumors, as they are the first group of sarcomas for which a chromosome translocation has been characterized at the molecular level (1, 2). This translocation, t(11;22), leads to the fusion of the EWS gene at 22q12, encoding for a putative RNA binding protein, to FLI1 at 11q24, an ETS-related gene coding for a transcription factor. Different breakpoints occur with in-frame fusion of EWS exons 7–10 to FLI1 exons 4–9 (3). The two most common combinations are EWS exon 7/ FLI1 exon 6 (termed type 1 fusion) and EWS exon 7/ FLI1 exon 5 (type 2 fusion). The biologic relevance of the different breakpoints is indicated by two retrospective analyses demonstrating that patients with nonmetastatic disease and a type 1 fusion had a better clinical outcome than patients with other exon combinations (4, 5). In another 5%–10% of Ewing tumor patients, EWS is fused to others members of the ETS family, most frequently ERG at 21q22 (6) or more rarely ETV1, E1AF and FEV(7–10). One of the target genes induced by EWS/FLI1 is manic fringe, a gene involved in somatic development. Overexpression of manic fringe by itself transforms NIH 3T3 cells (11). Another effect of EWS/FLI1 is suppression of the putative tumor suppressor gene transforming growth factor (TGF)-beta receptor II (12).
Despite these advances in the understanding of genetic aberrations and consecutive gene dysregulations, we still do not understand the biologic organization of the tumor, especially the processes that govern proliferation and differentiation of in vivo clonogenic tumor stem cells. For a long time, it has been known that only a portion of the cells in a tumor are clonogenic in vitro(13). However, as has been shown for normal and leukemic hematopoietic stem cells (14–16), clonogenic assays do not detect the most primitive cells and thus the relevance of these in vitro assays has been questioned. An in vivo model is, therefore, needed that allows detection and analysis of a primitive stem cell that can recapitulate the original disease. Such an in vivo model will also be a valuable tool to study many different biologic mechanisms involved in tumor metastasis, including organ-selective homing and tumor angiogenesis, and to develop and test new therapies.
So far, the majority of xenograft models for human solid tumors, including Ewing's sarcoma (17), have relied on subcutaneous propagation of tumor cells in athymic nude mice. However, this growth pattern does not recapitulate tumor cell growth and progression in patients. Moreover, because of the residual immune system in nude mice, not all tumor cell lines grow, and it has usually been impossible to engraft primary tumor cells. Therefore, nude mouse models, due to their lack of sensitivity, have been of limited value for human stem cell biology. Similar to the improvement of xenograft models for human hematopoietic stem cells (18), an important milestone in the development of xenograft models for human solid tumors has been made possible by the use of more immune-deficient mouse strains such as SCID mice (19–22). SCID mice lack functional B and T lymphocytes as they are unable to rearrange their immunoglobulin and T cell receptor genes. The latest improvement in the development of immune-deficient mouse strains are NOD/ scid mice. By crossing the scid mutation onto the NOD background, a mouse strain with a complex immune-deficiency involving B and T lymphocytes, NK cells, macrophages, and the complement system was created (23). These mice, by their almost complete inability to reject human xenotransplants, provide a more permissive environment for human cells than traditional nude mice.
In the present study, a novel approach was chosen to develop a biologically relevant xenograft model for human Ewing tumors. The natural course of untreated Ewing tumors leads to hematogenic metastasis in more than 90% of patients (24). Therefore, single-cell suspensions of human Ewing tumor cell lines and primary tumor cells were transplanted via intravenous injection onto highly immune-deficient NOD/ scid mice. Using this approach, we demonstrate that not only Ewing tumor cell lines but also primary Ewing tumor cells that are unable to proliferate as a cell line in vitro are able to engraft NOD/ scid mice. Transplanted mice develop a metastatic pattern similar to human patients including lung and bone tumors and a diffuse infiltration of the bone marrow. Analysis of integrins, i.e. adhesion molecules known to mediate homing and metastasis through cell-cell and cell-matrix interactions, showed that all Ewing tumor cell lines expressed the integrin α4β1 (VLA-4). α4β1 is known to be involved in targeting different cell types to the bone marrow and bone (25, 26). We have, therefore, been successful in establishing an in vivo model for human Ewing tumor cells that recapitulates many aspects of the growth and progression pattern of Ewing tumors in patients.
METHODS
Cell culture.
The human Ewing tumor cell lines VH-64, WE-68, and RM-82 were originally established by F.v.V. (27). VH-64 was isolated from a malignant pleural effusion (28) in a patient with Ewing's sarcoma of the metatarsal bone. WE-68 was cultured from a Ewing's sarcoma of the fibula with primary metastases to the lung (29). RM-82 was established from a Ewing's sarcoma of the femur with primary metastases to the tibia (29). TC-71 originates from a local Ewing's sarcoma relapse of the humerus (30) and was provided by Prof. T.J. Triche (Department of Pathology, Children's Hospital Los Angeles). CADO-ES1 cells (31) were purchased from the American Type Culture Collection (ATCC) (Rockville, MD, U.S.A.). CADO-ES1 cells were originally established from a malignant pleural effusion (31). These cell lines express the following EWS fusion transcripts: WE-68–EWS exon 7/ FLI1 exon 6 (type 1), VH-64 and TC-71–EWS exon 7/ FLI1 exon 5 (type 2), CADO-ES1 and RM-82–EWS exon 7/ ERG exon 6. The cells were propagated on collagen-coated tissue culture flasks in RPMI medium containing 10% FCS, 2 mM glutamine, and 100 U/mL penicillin and 100 μg/mL streptomycin at 37°C in a humidified atmosphere with 5% CO2. The cell lines were screened monthly for mycoplasma contamination using a DNA staining kit (Biochrom KG Seromed, Berlin, Germany).
Flow cytometric analysis of the expression of adhesion molecules.
Cultured TC-71, VH-64, WE-68, CADO-ES1, and RM-82 Ewing tumor cells were removed from collagen-coated culture flasks using 5 mM EDTA in PBS and washed twice with PBS at room temperature. A total of 5 × 105 cells in 20 μL aliquots were incubated for 15 min at room temperature at a concentration of 20 μg/mL of the following primary MAb: mouse anti-human integrin α1 (FB12), α2 (P1E6), α3 (P1B5), α5 (P1D6), rat anti-human integrin α6 (NKI-GoH3) (Chemicon, Temecula, CA, U.S.A.), mouse anti-human integrin α4 (44H6), and β1 (TDM29) (Dianova, Hamburg, Germany). Subsequently, cells were washed with PBS at room temperature and 20 μL of a 1:50 dilution of F(ab′)2 fragment goat anti-mouse IgG FITC/PE conjugates or anti-rat IgG DTAF conjugates were added (Dianova, Hamburg, Germany, and Immunotech, Marseille, France, respectively). After an incubation period of 15 min at room temperature, cells were washed again and each sample was resuspended after addition of 100 μL PBS. Isotype antibodies served as controls (mouse IgG1 and rat IgG2a, Immunotech, Marseille, France; mouse IgG3, Dianova, Hamburg, Germany). The cells were analyzed using a FACSCalibur flow cytometer and Cell-Quest software (Becton-Dickinson, Heidelberg, Germany).
Transplantation of human Ewing tumor cells onto immuno-deficient mice.
NOD/ scid mice (Jax® mice stock name: NOD/LtSz-Prkdcscid/J mice) were provided by Prof. John E. Dick (Toronto, Ontario, Canada), with permission from Dr. Len Shultz (Jackson Laboratories, Bar Harbor, ME, U.S.A.). The NOD/ scid mice (23) were bred and maintained under defined flora conditions in individually ventilated microisolator cages (Lab Products, Maywood, NJ, U.S.A.) at the University of Münster as previously described (32). In addition to the scid mutation, NOD/ scid mice have a very low natural killer cell activity, a deficiency in the C5 component of the complement system and a functional macrophage defect (23). Eight- to 16-wk-old NOD/ scid mice were irradiated with a single dose of 3.5 Gy from a cobalt 60-unit 1 d before transplantation. For transplantation, the tumor cells were recovered from the tissue culture flask using PBS buffer containing 5 mM EDTA. Into the tail vein of each mouse, 1–5 × 106 cells as single-cell suspension in 0.5 mL RPMI were injected. Fresh tumor samples were cut into small pieces under sterile conditions and carefully minced through 40-μm nylon cell strainers (Becton Dickinson). To exclude clumping of tumor cells, cell suspensions were microscopically checked before transplantation. Mice that died within 3 wk after transplantation due to transplant-related toxicity (n = 6), such as infections, and those that died unexpectedly (n = 3) had to be excluded from the analysis. Animals were killed immediately when they developed clinical signs of tumor development. The mice were carefully dissected and macroscopically screened for tumor formation. All experimental manipulations with the mice were performed under sterile conditions in a laminar flow hood. The animal experiments were approved by the animal care committee of the local government (Bezirksregierung Münster, Aktenzeichen 23.0835.1.0 (73/95)).
Light microscopic examination of transplanted mice.
Brain, lungs, kidneys, liver, and spleen were routinely examined in all mice. These organs and macroscopically detected tumors were fixed in 4% buffered formalin, processed, and embedded in paraffin. Three-micrometer sections were cut and stained with H&E and PAS.
Immunohistochemistry.
Four-micrometer sections were prepared from formalin-fixed and paraffin-embedded material and mounted onto silane-coated slides. The sections were stained for huCD99 with a MAb (O13, Signet Laboratories, Dedham, MA, U.S.A.) using a standard APAAP protocol.
RT-PCR for EWS/FLI1 and EWS/ERG fusion transcripts.
To detect human Ewing tumor cells in transplanted mice, RNA was extracted from suspected tumors and the murine bone marrow. Total RNA was converted to cDNA using a first strand cDNA synthesis kit according to the manufacturer's protocol (Pharmacia Biotech, Freiburg, Germany). In this procedure an oligo(dT) primer and a FLI1/ERG sequence-specific primer (5′GAAGGGTACTTGTACA3′) were used for generating the first strand cDNA in a final reaction mix of 15 μL. A 1–4 μL sample of the first strand synthesis reaction was used for amplification in a 50-μL standard PCR reaction employing the primers A1 5′CCA CTA GTT ACC CAC CCC AAA C3′ (EWS) and A2 5′AAC TCC CCG TTG GTS CCY TCC3′ (consensus oligonucleotide FLI1/ERG). A “nested PCR” was added using the internal primers B1 5′TCC TAC AGC CAA GCT CCA AGT C3′ (EWS(1),) and B2 5′GAA TTG CCA CAG CTG GAT CTG C3′ (FLI1) or B1 and B3 5′GAG TTG GAG CTG TCC GAC AGG3′ (ERG), respectively (33). Amplification was achieved by 30 cycles with denaturation at 94°C for 30 s, annealing of primers at 65°C for 30 s, and extending at 72°C for 45 s. The suitability of the extracted RNA was tested by reverse-transcriptase PCR (RT-PCR) amplification of a 495 bp fragment of the human β-actin cDNA. Negative controls were included using RNA from cytogenetically t(11;22)-negative cell lines.
Flow cytometric analysis of the murine bone marrow.
To quantify and characterize human Ewing tumor cells infiltrating the murine marrow, two- and three-color flow cytometry was performed according to standard procedures (34, 35). In short, bone marrow cells were incubated with saturating amounts of FITC-, PE-, and CyChrome-conjugated antibodies for 20 min at room temperature. Subsequently, the erythrocytes were lysed (FACS Lysing Solution, Becton Dickinson, Heidelberg, Germany) and the cells analyzed on a FACSCalibur (Becton Dickinson). Human cell were detected by anti-huHLA-ABC-FITC (B9.12.1, Immunotech, Hamburg, Germany), Ewing tumor cells by anti-huCD99 (MIC-2) (TÜ 12, PharMingen, Hamburg, Germany) and murine hematopoietic cells by anti-muCD45-CyChrome (30-F11, PharMingen). Matched isotype controls were included in each experiment. As the anti-huHLA-ABC and anti-huCD99 antibodies, unlike the isotype controls, produce low levels of background staining in the murine bone marrow (most likely due to cross-reactivity with murine antigens) anti-muCD45 antibody was used to gate out murine hematopoietic cells. Data acquisition and analysis was performed with Cell-Quest and Paint-a-Gate software (Becton Dickinson).
X-ray analysis.
X-ray analysis of killed mice was performed using a FeinFocus x-ray machine. Two standard x-ray films (Kodak X-OMAT, Kodak, Stuttgart, Germany) were exposed at 25–30 KV for 7 and 9 s and automatically developed.
Quantitation of the engraftment unit.
The statistical analysis is based on the assumption that only a fraction of the Ewing tumor cells is capable of engrafting the immune-deficient mice and that a single cell, i.e. one engraftment unit, is sufficient to reestablish tumor growth in transplanted animals. In analogy to the hematopoietic system, we operationally call this engraftment unit ET-IC. Pooled data from the limiting dilution experiments with VH-64 cells were analyzed with a five-digit accuracy (36). ET-IC, variance, SE, and 95% confidence interval were determined by maximum likelihood and minimum χ2 estimation. Both maximum likelihood and minimum χ2 estimation are based on a Poisson probability distribution and estimate the probability that y of x transplanted mice develop a tumor. These estimators are designed to calculate the fraction of ET-IC in a manner so that the results observed are the most probable according to mathematical statistics. If the estimations yielded by maximum likelihood and minimum χ2 estimation do not differ more than 10%, the results from the minimum χ2 estimation are accepted.
RESULTS
Engraftment and metastasis of Ewing tumor cell lines in immune-deficient NOD/ scid mice.
To establish an in vivo metastasis model for human Ewing tumors, 0.1–5 × 106 cells from the Ewing tumor cell lines VH-64, WE-68, CADO-ES1, TC-71, and RM-82 were transplanted via intravenous injection into immune-deficient NOD/ scid mice. Overall, 26 of 52 mice (50%) transplanted with these cell lines developed macroscopic tumors (Table 1A).
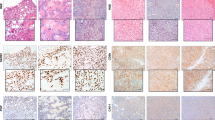
The initial experiments were carried out with VH-64 cells. Thirteen out of 27 mice (48%) transplanted with 0.1–5 × 106 VH-64 cells developed tumors within 22–160 d after injection (Table 1A). On dissection and histologic and molecular analysis, engrafted mice had lung (77% of engrafted mice) (Fig. 1C), kidney (46%) (Fig. 1E), bone (31%) (Fig. 1A) and subcutaneous tumors (8%), and an infiltration of the bone marrow (44%) (Table 1B). In addition, female mice developed ovarian metastases at high frequency (80%). Similar results were observed after transplantation of 2–5 × 106 WE-68, CADO-ES1, TC-71, and RM-82 cells (Table 1A/B). Overall, 62% of engrafted mice developed lung, 46% kidney, 23% bone, 11% subcutaneous metastases and 30% a bone marrow infiltration as detected by RT-PCR (Table 1B). On H&E staining of the different tissue sections, the Ewing tumors in the mice showed the characteristic small blue round cell morphology typical for this tumor entity (Fig. 1,A–C and E). The bone marrow infiltration was assessed by RT-PCR analysis of EWS fusion transcripts in RNA extracted from the murine bone marrow as described in “Methods” and the legend of Figure 2. Mice #2.7.1 and #4.16.3 are presented as typical examples (Fig. 2). A RT-PCR product of the same size could be amplified from RNA extracted from the original Ewing tumor cell lines (VH-64 and WE-68) and the bone marrow of the transplanted mice (Fig. 2A). The specificity of the RT-PCR products is demonstrated by hybridization with EWS exon 7- and FLI-1 exon 8-specific probes (Fig. 2B). The tumors were highly vascularized demonstrating that human Ewing tumor cells were able to induce angiogenesis in the murine environment (data not shown).
Light microscopic and immunohistochemical examination of transplanted NOD/ scid mice. (A) H&E-stained small blue round cell tumor of the femur from mouse #2.7.1, transplanted with 1 × 106 VH-64 cells (Tu, tumor tissue; B, normal bone). (B) H&E-stained lung metastasis from mouse #9.25.1, transplanted with 5 × 106 tumor cells from patient UPN #2 (Met, metastasis;Lu, normal lung). (C) Two H&E-stained lung metastases from mouse #2.30.1, transplanted with 2 × 106 VH-64 cells. (D) Typical tumor cell membrane staining for huCD99 (red, APAAP method) of a lung metastasis from mouse #2.30.1. (E) H&E-stained kidney tumor from mouse #3.30.4 transplanted with 5 × 106 VH-64 cells (Tu, tumor tissue;Kid, normal kidney). (F) huCD99 staining (red) of the kidney tumor from mouse #3.30.4.
Detection of EWS fusion transcripts. (A) RT-PCR for EWS fusion transcripts in samples derived from engrafted mice. Using EWS/FLI1, respectively EWS/ERG specific primer pairs, the same PCR product could be generated in the transplanted Ewing tumor cells and tissues from engrafted NOD/ scid mice: in the cell line VH-64 and in a tumor (Tumor) of the femur and the bone marrow (BM) of mouse #2.7.1, transplanted with 1 × 106 VH-64 cells; in the cell line WE-68 and a subcutaneous neck tumor and the bone marrow of mouse #4.16.3, transplanted with 2 × 106 WE-68 cells; in the cell line CADO-ES1 and two bone tumors (shoulder and rip cage) but not the bone marrow of mouse #5.29.7; the primary scapula tumor of patient #1 (UPN1) and in the subcutaneous and lung tumor of mouse #8.26.2, transplanted with 6 × 106 cells from this patient. Hae III-digested PhiX 174 DNA was included as a size marker. DNA was separated by nondenaturing PAGE and detected by silver staining. (B) The specificity of the amplified fragments for the translocation junctions was confirmed by Southern blotting of the amplified DNA using oligonucleotides corresponding to EWS exon 7, FLI1 exon 8, and ERG exon 8 as hybridization probes. Samples derived from cell lines VH-64, WE-68, and the tumor from patient #1 show specific signals using EWS and FLI1 oligonucleotides indicating a t(11;22)-translocation. In the CADO-ES1 cell line and the subcutaneous and lung tumors in mouse #5.29.7, ERG is fused to EWS corresponding to a t(21;22)-translocation.
Immunohistochemical and molecular identification of human Ewing tumor cells.
To confirm that the tumors in the mice originated from the transplanted Ewing tumor cells, histologic sections of different tissues were stained for H&E, PAS, and huCD99, an antigen known to be expressed on Ewing tumors. Lung and bone metastases were PAS-positive (data not shown) and clearly expressed huCD99 as seen by the red APAAP staining of the tumor cell membranes (Fig. 1D). The same membrane-bound expression of huCD99 was seen in metastases in other organs, such as ovarian, kidney (Fig. 1F), and subcutaneous metastases.
In addition, RNA was extracted from the different organs and individual tumor nodules. RT-PCR analysis for EWS-fusion transcripts detected the same EWS/FLI1, respectively EWS/ERG fusion transcripts in RNA extracted from the tumors and the bone marrow of transplanted mice and the transplanted cell lines. A representative analysis of six tumors from four different mice (mouse #2.7.1 transplanted with VH-64 cells, mouse #4.16.3 transplanted with WE-68 cells, mouse #5.29.7 transplanted with CADO-ES1 cells, and mouse #8.26.2 transplanted with primary tumor cells from patient UPN#1) is shown in Figure 2. RT-PCR products of the same size could be amplified from RNA extracted from the original Ewing tumor cell lines (VH-64, WE-68, CADO-ES1) or the primary tumor from patient UPN#1 and RNA extracted from tumors of the transplanted mice (Fig. 2A). The specificity of the RT-PCR products was demonstrated by hybridization with EWS exon 7- and FLI-1 exon 8-, respectively. ERG exon 8-specific probes (Fig. 2B). These results prove that human Ewing tumor cells engraft and proliferate in NOD/ scid mice.
Bone marrow infiltration and bone tumor formation.
One of the characteristic features of Ewing tumors is the high frequency of bone marrow involvement. Consistent with this clinical observation, 6 out of 20 tumor-bearing mice (30%) from which high-quality RNA was available showed a bone marrow involvement as detected by RT-PCR for EWS-fusion transcripts (Table 1B and Fig. 2). The presence of human cells in the murine marrow was also confirmed by Southern blot analysis and genomic PCR specific for human muscular dystrophy gene sequences (data not shown). In the marrow of two of these six mice, the levels of human cells in the murine marrow were high enough (0.6%–3.5%) to be detected by flow cytometry. For flow cytometry, single-cell suspensions of the transplanted Ewing tumor cell line (positive control), bone marrow from transplanted mice and bone marrow from untransplanted mice (negative control) were incubated with FITC-labeled antibodies recognizing human HLA class I molecules, PE-labeled antibodies binding to human CD99, and CyChrome-labeled antibodies detecting murine CD45 (muCD45) and subsequently analyzed with a flow cytometer as described in “Methods.” The analysis of the bone marrow from mouse #3.30.4 transplanted with VH-64 Ewing tumor cells is shown in Figure 3. To reduce background, the majority of murine hematopoietic cells were excluded from the analysis by gating (gate R1) for cells with absent of low expression of murine CD45. Using this gating strategy, a small population of human Ewing tumor cells (0.6% of all analyzed events) that had a comparable expression pattern for human class I antigens and huCD99 as the transplanted cell line could be detected in the bone marrow of this mouse. Thus, these results show that human Ewing tumor cells cause an infiltration in murine marrow similar to that seen in patients' marrow.
Flow cytometric analysis of bone marrow-infiltrating Ewing tumor cells in NOD/ scid mice. Upper two plots : analysis of the original cell line VH-64. Middle two plots : the bone marrow of mouse #3.30.4, that was transplanted with 5 × 106 VH-64 cells. Lower two plots : the bone marrow of a nontransplanted control mouse. Histograms on the left : for this analysis, a gate (R1) was set on all events that were negative for murine CD45 (muCD45 CyChrome). Dot plots on the right : the events in the right upper quadrant represent huCD99+HLA-ABC+ human Ewing tumor cells.
In correlation with the frequent tumor infiltration of the murine marrow, a significant percentage of transplanted mice also developed bone tumors, i.e. 6 of 26 (23%) of tumor-bearing mice presented with bone metastases (Table 1B). This was proven by histology (Fig. 1A), immunohistochemistry, and RT-PCR (Fig. 2). Moreover, x-ray analysis of one of these bone tumors demonstrated a characteristic appearance with osteolytic lesions, destruction of the cortex, and infiltration of the surrounding soft tissue (Fig. 4).
Pattern of metastasis and integrin expression.
Several observations suggested that the pattern of metastasis of human Ewing tumor cells in the NOD/ scid mice was tumor- and not host-specific: the high frequencies of lung and bone tumors and of bone marrow infiltration were similar to those seen in patients. No metastasis to other organs such as liver, spleen and brain could be detected in any tumor-bearing mice, and the pattern of metastasis in the immune-deficient mice was different to what has been described for other tumor cell types, such as neuroblastoma (19).
Organ-selective homing and metastasis is known to be mediated by adhesion molecules, particularly β1-integrins (VLA family) (37). Integrins are heterodimeric molecules that are composed of α- and β-chains. Expression of the β1-integrins was analyzed on the transplanted Ewing tumor cell lines (Fig. 5) and was correlated with the pattern of metastasis. Interestingly, and in correlation with the high frequency of bone marrow infiltration and the formation of bone tumors, all Ewing tumor cell lines expressed α4β1 (VLA-4), an integrin that is involved in targeting different cell types to bone and bone marrow (25, 26). A similar expression pattern was seen for the fibronectin receptor α5β1 (VLA-5), an integrin that is expressed on primitive hematopoietic stem cells and thought to be important for the interaction of hematopoietic stem cells with the bone marrow microenvironment (38). With the exception of CADO-ES1 cells, all Ewing tumor cell lines expressed the collagen receptor α2β1, whereas a heterogeneous expression was detected for the integrin α1β1 and only low to zero expression for the integrins α3β1 and α6β1 (Fig. 5).
Expression of VLA integrin subunits on Ewing tumor cell lines analyzed by flow cytometry. TC-71, VH-64, WE-68, CADO-ES1, and RM-82 Ewing tumor cells were stained with anti-α1 (FB12), -α2 (P1E6), -α3 (P1B5), -α4 (44H6), -α5 (P1D6), -α6 (NKI-GoH3), and -β1 (TDM29) MAb and respective FITC/DTAF/PE labeled secondary F(ab′)2 fragments. Representative fluorescence histograms from 4 to 12 independent experiments are shown (x axis, log fluorescence intensity;y axis, relative cell number;black graphs, integrin subunit expression;gray graphs, respective isotype controls).
To facilitate comparisons of different cell lines, higher numbers of mice were transplanted with VH-64 and CADO-ES1 cells. These two cell lines differ in many aspects. VH-64 carry the classical t(11;22) with an EWS-FLI1 fusion transcript while CADO-ES1 has a variant t(21;22) with an EWS-ERG fusion transcript. Moreover, both cell lines differ in their expression of β1-integrins as CADO-ES1 unlike VH-64 cells lack α1β1 and α2β1. Interestingly, both cell lines also show a different pattern of metastasis with CADO-ES1-transplanted mice having fewer lung tumors, no detectable bone marrow infiltration, but a higher frequency of tumors in internal organs.
This pattern of integrin expression may be involved in organ-selective metastasis of Ewing tumor cells in NOD/ scid mice as well as in patients. Therefore, tumor cell homing, including the role of different integrins, can now be studied in this Ewing tumor-NOD/ scid mouse model.
Quantitative analysis of the engraftment unit.
To test if the described transplantation assay represents a quantitative assay for in vivo clonogenic tumor cells capable of reestablishing the disease in mice, VH-64 cells were transplanted at increasing cell doses. Rates of engraftment were as follows: none of 3 mice (0%) transplanted with 0.1 × 106 cells, 4 of 14 mice (29%) transplanted with 1–2 × 106 cells, and 9 of 10 mice (90%) transplanted with 5 × 106 cells (Table 1A). Time to engraftment was also cell dose-dependent with mice transplanted at the higher cell dose of 5 × 106 developing macroscopic disease faster, i.e. within 22–49 d, compared with mice transplanted with 1–2 × 106 tumor cells, i.e. within 36–160 d.
Pooled data from these limiting dilution experiments were statistically analyzed according to standard methods (36). In short, minimal χ2 estimation was used to calculate the frequency of the engraftment unit, i.e. the frequency of the cell engrafting in the transplanted animals. This engraftment unit is operationally called ET-IC. Maximum likelihood estimation is only applied as a statistical control method, and if the estimations yielded by maximum likelihood and minimum χ2 estimation do not differ by more than 10%, the results from the minimum χ2 estimation are accepted.
The minimal χ2 estimation of the above data calculated a fraction of ET-IC of 3.2019 × 10–7 in the total population of transplanted VH-64 cells (i.e. 1 ET-IC in 3.123 × 106 tumor cells) with a SE of 1.0503 × 10–7 and a 95% confidence interval of 1.1433–5.2606 × 10–7 (1 ET-IC in 1.905–8.747 × 106 tumor cells). As the results of the maximum likelihood estimation (ET-IC fraction of 3.2771 × 10–7, i.e. 1 ET-IC in 3.051 × 106 VH-64 cells) differed only by 2.3% the results from the minimal χ2 estimation were accepted. Thus, by statistical analysis, only 1 cell out of 3 × 106 transplanted VH-64 tumor cells engrafted the mice.
Engraftment of primary tumor cells.
Cell lines may not reflect all aspects of primary tumors as they have acquired many additional genetic changes during immortalization and during cell culture over time. This is also exemplified by the observation that patients from whom a cell line can be established have a dismal prognosis (27). These difficulties to grow primary tumor cells in vitro demonstrate that the signals, such as growth factors, stromal elements and cell-cell interactions, that Ewing tumor cells need to survive and proliferate are poorly understood. Because of the importance to study primary tumor cells it was tested if the in vivo microenvironment in NOD/ scid mice is able to support growth and survival of primary Ewing tumor cells. Single-cell suspensions from tumors of three different patients were transplanted into NOD/ scid mice. Transplantation of 4–6 × 106 cells of two of the patients (UPN #1 and #2) led to tumor formation in four out of seven mice (57%) (Table 1A). The first patient (UPN #1) had a primary scapula tumor and isolated lung metastases. The tumor material was taken from the initial diagnostic biopsy of the scapula tumor. By molecular analysis, the tumor cells were shown to carry a type 1 EWS/FLI1 fusion transcript. Compared with the experiments with cell lines in which tumor formation in transplanted mice occurred after 1–6 mo, the primary tumor cells needed 4–9 mo to cause macroscopic disease. During this time interval, the mice developed lung and subcutaneous metastases. As in the patient, no infiltration of the murine marrow could be detected by RT-PCR analysis (Fig. 2). Interestingly, primary cells from the same sample did not grow as a cell line in vitro. Cells from the second patient (UPN #2) with a local Ewing tumor relapse led to the development of a lung metastasis (Fig. 1B) in one out of three transplanted mice. These cells also grew as a cell line in vitro. Cells from the third patient (UPN #3) did not engraft. Although based on small numbers of patients due to the limited availability of primary tumor samples, these data prove that the in vivo microenvironment in NOD/ scid mice is more permissive for the survival and growth of primary Ewing tumor cells than current culture conditions in vitro.
DISCUSSION
In this study we have been able to establish a novel in vivo model for human Ewing tumor cells that recapitulates many aspects of growth and progression of Ewing tumors in patients. This xenograft model is based on two important improvements compared with classical nude mouse models for solid tumors. First, and in accordance with the hematogenic spread of Ewing tumors, single-cell suspensions of the tumor were transplanted via intravenous injection. Secondly, an improved mouse strain, i.e. NOD/ scid mice, was used as recipient that nearly completely lacks the ability to reject human cells. NOD/ scid mice carry a complex immune deficiency involving B and T lymphocytes, NK cells, macrophages, and the complement system (23).
In accordance with the profound immune deficiency of the recipient mice, primary Ewing tumor cells engrafted. More importantly, tumor cells from patient UPN#1 that engrafted the mice did not grow in vitro. This is of particular importance as only few primary Ewing tumors grow in vitro. The ability to generate a cell line from a patient correlates with a dismal prognosis. This indicates that Ewing tumor cells need additional genetic alterations to grow in vitro that are different from the primary events leading to the malignant transformation. Thus, as has been shown for neuroblastoma (19) and prostate cancer cells (20), these data confirm that immune deficient scid and NOD/ scid mice offer a suitable microenvironment for primary human tumor cells that cannot be provided by current in vitro assays.
A small fraction of primitive tumor stem cells is assumed to maintain the malignant cell clone in vivo(13, 39), however, so far no suitable in vivo assays for solid tumor stem cells have been available. Therefore, to characterize and quantitate those Ewing tumor cells capable of engrafting and proliferating in immune-deficient mice, NOD/ scid mice were transplanted with increasing cell doses of VH-64. Our experiments established that Ewing tumor cell engraftment was cell dose-dependent (Table 1A) and the frequency of the engraftment unit, termed the ET-IC, was 1 in 3 × 106 VH-64 cells. However, this statistical analysis underestimates the actual frequency of ET-IC as not every cell that is capable of engrafting the mice engrafts. Quantitative analysis has shown that the seeding efficiency of primitive human hematopoietic cells in NOD/ scid mice in different organs varies between 2.3% and 8.7%(40). Assuming a similar seeding efficiency for solid tumor cells of 5%–10%, this means that approximately 1 in 1.5–3 × 105 VH-64 Ewing tumor cells is capable of engrafting and proliferating in NOD/ scid mice.
As the clonogenic frequency in vivo was significantly lower than the clonogenic frequency in vitro (57% ± 6.5% of VH-64 cells from spheroid colonies in a methylcellulose assay), these data are compatible with the hypothesis that a similar hierarchy exists within the malignant cell clone of solid tumors as has been characterized for leukemias (14, 15, 41) : most Ewing tumor cells have a limited proliferative capacity whereas only a small population of malignant stem cells with a basically indefinite proliferative and self-renewal capacity maintains the malignant cell clone in vivo. According to this stem cell concept of solid tumors, the malignant transformation occurs in a normal tissue stem cell responsible for tissue regeneration. Ewing tumors, in particular, are thought to arise from an embryonic stem cell, which is hypothesized to be of neuroectodermal origin (42, 43) but has yet to be identified. Although still controversial, this concept of a ubiquitous pluripotent embryonic neuroectodermal stem cell may explain the heterogeneity in Ewing tumor differentiation, i.e. peripheral neuroectodermal tumors versus classical Ewing's sarcoma, and the heterogeneity in localization, i.e. bone versus extraskeletal tumors. These open questions concerning the origin of embryonic mesenchymal tumors highlight the importance of an in vivo model for solid tumor stem cell biology.
Engrafted mice developed lung, kidney, ovary, bone, and subcutaneous tumors as well as a diffuse bone marrow infiltration (Table 1A). The formation of bone tumors and bone marrow infiltration together with the high frequency of lung metastases resembles many aspects of disease progression in patients. The time frame of tumor formation in the mice was also comparable to that observed in human patients. Cell lines took 1–6 mo and primary tumor cells 4–9 mo to cause macroscopic disease in transplanted mice. In patients, relapse usually occurs within 1 y after completion of therapy (44), indicating that low numbers of Ewing tumor cells escaping primary therapy need several months to grow up to clinically detectable metastases.
In contrast to the clinical setting, Ewing tumor cells transplanted into immune-deficient NOD/ scid mice more frequently metastasize to kidneys and ovaries. In human patients, metastases of the kidneys are rare, but with more widespread use of immunohistochemistry and molecular techniques for pathologic diagnosis, primary Ewing tumors of the kidney are increasingly diagnosed (45–47). Human tumor metastases to murine ovaries appears to be a common phenomenon (19) and may be host specific. However, Ewing tumor metastases to the ovaries have also been described in individual patients (48–50). Thus, the overall growth and metastasis pattern in the mice is surprisingly similar to that in patients. In addition, and also in accordance with naturally occurring Ewing tumors, no metastases to liver, spleen, or brain were detected in any transplanted mice.
Tumor cell metastasis is a complex process. The described Ewing tumor-NOD/ scid mouse model represents the late stages of metastasis after the tumor cells have managed to disaggregate from the primary tumor and invade the blood stream. These late stages of tumor cell metastasis include tumor cell adhesion to the endothelial wall, transendothelial migration and tissue invasion (chemotaxis), the proliferative and self-renewal potential of individual tumor cells, and induction of angiogenesis.
To gain insight into the mechanisms of organ-selective metastasis of Ewing tumor cells in vivo, expression of integrins was analyzed on the transplanted cell lines. Integrins are involved in cell-cell and cell-matrix interactions and play a central role in homing and metastasis. It has been demonstrated that Chinese hamster ovary (CHO) cells transfected with the integrin α4β1 (VLA-4), unlike parental CHO cells, metastasize to the murine bone (26). α4β1 has also been shown to be involved in homing of hematopoietic stem cell to the bone marrow (25). In accordance with these published results and with the high frequency of bone marrow and bone metastasis in the mice, all Ewing tumor cell lines expressed α4β1. All Ewing tumor cell lines also expressed the fibronectin receptor α5β1 (VLA-5), which is thought to be involved in the interaction of hematopoietic stem cells with the bone marrow microenvironment (38). In vitro studies demonstrated that α4β1 and α5β1 are functional and mediate adhesion and migration of Ewing tumor cells to endothelial VCAM-1 and the extracellular matrix component fibronectin, respectively (Decker et al., manuscript in preparation). Thus, Ewing tumor cells may use similar mechanisms as primitive hematopoietic cells for homing to the bone marrow. In addition to the described adhesion molecules that mediate interaction of the tumor cells with the endothelial wall and extracellular matrix, we have also been able to show that Ewing tumor cells express the chemokine receptor CXCR4 and the angiogenic vascular endothelial growth factor (VEGF) (Hotfilder et al., unpublished results). For the first time, this Ewing tumor-NOD/ scid mouse model provides a tool to study the complex interactions of metastasizing Ewing tumor cells with the microenvironment of the target tissue in vivo, including tumor cell homing, migration, growth, and angiogenesis.
Finally, tumor cells with a wide spectrum of different EWS fusion transcripts engrafted. Interestingly, VH-64 cells with a EWS exon 7/ FLI1 exon 5 (type 2) fusion transcript produced a different pattern of metastasis in the mice as compared with CADO-ES1 cells that carry an EWS exon 7/ ERG exon 6 fusion transcript. This may be explained by expression of different integrins (see above) and/or differences in the underlying genetic aberrations. As clinical trials suggested a biologic relevance of different EWS fusion transcripts, an in vivo model such as the described Ewing tumor-NOD/ scid mouse model will be fundamental to elucidate the role of these different genetic aberrations.
In conclusion, using severely immune-deficient NOD/ scid mice as recipients and choosing an intravenous transplantation approach, it has been possible to establish an in vivo model that recapitulates many aspects of growth and progression of human Ewing tumors.
Abbreviations
- APAAP method:
-
alkaline phosphatase anti-alkaline phosphatase method
- CyChrome:
-
phycoerythrin-cyanine
- DTAF:
-
dichlorotrianzinylamino-fluorescein
- ET-IC:
-
Ewing tumor-initiating cell
- H&E:
-
hematoxylin and eosin
- HLA:
-
human leukocyte antigen
- NOD/ scid mice:
-
nonobese diabetes/severe combined immune-deficiency mice
- PAS:
-
periodic acid-Schiff reaction
- PE:
-
phycoerythrin
- PNET:
-
peripheral neuroectodermal tumor
- VCAM:
-
vascular adhesion molecule
- VLA:
-
very late activation antigen
References
Delattre O, Zucman J, Plougastel B, Desmaze C, Melot T, Peter M, Kovar H, Joubert I, de Jong P, Rouleau G, Aurias A, Thomas G 1992 Gene fusion with an ETS DNA-binding domain caused by chromosome translocation in human tumours. Nature 359: 162–165
Delattre O, Zucman J, Melot T, Garau XS, Zucker JM, Lenoir GM, Ambros PF, Sheer D, Turc-Carel C, Triche TJ, Aurias A, Gilless T 1994 The Ewing family of tumors—a subgroup of small-round-cell tumors defined by specific chimeric transcript. N Engl J Med 331: 325–327
Zucman J, Melot T, Desmaze C, Ghysdael J, Plougastel B, Peter M, Zucker JM, Sheer TJ, Turc-Carel C, Ambros P, Combaret V, Lenoir G, Aurias A, Thomas G, Delattre O 1993 Combinatorial generation of variable fusion proteins in the Ewing family of tumours. EMBO J 12: 4481–4487
de Alava E, Kawai A, Healey JH, Fligman I, Meyers PA, Huvos AG, Gerald WL, Jhanwar SC, Argani P, Antonescu CR, Pardo-Mindan FJ, Ginsberg J, Womer R, Lawlor ER, Wunder J, Andrulis I, Sorensen PHB, Barr FG, Ladanyi M 1998 EWS-FLI1 fusion transcript structure is an independent determinant of prognosis in Ewing's sarcoma. J Clin Oncol 16: 1248–1255
Zoubek A, Dockhorn-Dworniczak B, Delattre O, Christiansen H, Niggli F, Gatterer-Menz I, Smith TL, Jürgens H, Gadner H, Kovar H 1996 Does expression of different EWS chimeric transcripts define clinically distinct risk groups of Ewing tumor patients?. J Clin Oncol 14: 1245–1251
Sorensen PHB, Lessnick SL, Lopez-Terrada D, Liu XF, Triche TJ, Denny CT 1994 A second Ewing's sarcoma translocation, t(21;22), fuses the EWS gene to another ETS-family transciption factor, ERG. Nat Genet 6: 146–151
Jeon I-S, Davis JN, Braun BS, Sublett JE, Roussel MF, Denny CT, Shapiro DN 1995 A variant Ewing's sarcoma translocation (7;22) fuses the EWS gene to the ETS gene ETV1. Oncogene 10: 1229–1234
Kaneko Y, Yoshida K, Handa M, Toyoda Y, Nishihira H, Tanaka Y, Sasaki Y, Ishida S, Higashino F, Fujinaga K 1996 Fusion of an ETS-family gene, E1AF, to EWS by t(17;22)(q12;q12) chromosome translocation in an undifferentiated sarcoma of infancy. Genes Chromosomes Cancer 15: 115–121
Peter M, Couturier J, Pacquement H, Michon J, Thomas G, Magdelenat H, Delattre O 1997 A new member of the ETS family fused to EWS in Ewing tumors. Oncogene 14: 1159–1164
Urano F, Umezawa A, Hong W, Kikuchi H, Hata J 1996 A novel chimera gene between EWS and E1AF, encoding the adenovirus E1A enhancer-binding protein, in extraosseous Ewing's sarcoma. Biochem Biophys Res Commun 219: 608–612
May WA, Arvand A, Thompson AD, Braun BS, Wright M, Denny CT 1997 EWS-FLI1-induced manic fringe renders NIH 3T3 cells tumorigenic. Nat Genet 17: 495–497
Hahm KB, Cho K, Lee C, Im YH, Chang J, Choi SG, Sorensen PH, Thiele CJ, Kim SJ 1999 Repression of the gene encoding the TGF-beta type II receptor is a major target of the EWS-FLI1 oncoprotein. Nat Genet 23: 222–227
Selby P, Buick RN, Tannock I 1983 A critical appraisal of the human tumour stem-cell assay. N Engl J Med 308: 129–134
Bonnet D, Dick JE 1997 Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat Med 3: 730–737
Lapidot T, Sirard C, Vormoor J, Murdoch B, Hoang T, Caceres-Cortes J, Minden M, Patterson B, Caligiuri MA, Dick JE 1994 A cell initiating human acute myeloid leukaemia after transplantation into SCID mice. Nature 367: 645–648
Cashman JD, Lapidot T, Wang JC, Doedens M, Shultz LD, Lansdorp P, Dick JE, Eaves CJ 1997 Kinetic evidence of the regeneration of multilineage hematopoiesis from primitive cells in normal human bone marrow transplanted into immunodeficient mice. Blood 89: 4307–4316
Scotlandi K, Benini S, Nanni P, Lollini P-L, Nicoletti G, Landuzzi L, Serra M, Manara MC, Picci P, Baldini N 1998 Blockage of insulin-like growth factor-I receptor inhibits the growth of Ewing's sarcoma in athymic mice. Cancer Res 58: 4127–4131
Vormoor J, Ritter J, Jürgens H, Dick JE 1995 Significance of the SCID mouse models for the study of normal and leukemic human hemopoiesis and gene therapy. Onkologie 18: 434–438
Bogenmann E 1996 A metastatic neuroblastoma model in SCID mice. Int J Cancer 67: 379–385
Klein KA, Reiter RE, Redula J, Moradi H, Zhu XL, Brothman AR, Lamb DJ, Macelli M, Belldegrun A, Witte ON, Sawyers CL 1997 Progression of metastatic human prostate cancer to androgen independence in immunodeficient SCID mice. Nature Med 3: 402–408
Phillips RA, Jewett MAS, Gallie BL 1989 Growth of human tumors in immune-deficient scid mice and nude mice. Curr Top Microbiol Immunol 152: 259–263
Visonneau S, Cesano A, Torosian MH, Miller EJ, Santoli D 1998 Growth characteristics and metastatic properties of human breast cancer xenografts in immunodeficient mice. Am J Pathol 152: 1299–1311
Shultz LD, Schweitzer P, Christianson S, Gott B, Birdsall-Maller I, Tennent B, McKenna S, Mobraten L, Rajan T, Greiner D, Leiter E 1995 Multiple defects in innate and adaptive immunological function in NOD/LtSz-scid mice. J Immunol 154: 180–191
Ewing J 1921 Diffuse endothelioma of bone. Proc N Y Pathol Soc 21: 17–24
Quesenberry PJ, Becker PS 1998 Stem cell homing: rolling, crawling, and nesting. Proc Natl Acad Sci U S A 95: 15155–15157
Matsuura N, Puzon-McLaughlin W, Irie A, Morikawa Y, Kakudo K, Takada Y 1996 Induction of experimental bone metastasis in mice by transfection of integrin α4β1 into tumor cells. Am J Pathol 148: 55–61
van Valen F 1999 Ewing's sarcoma family of tumors. Hum Cell Cult 1: 55–85
van Valen F, Winkelmann W, Jürgens H 1992 Type I and type II insulin-like growth factor receptors and their function in human Ewing's sarcoma cells. J Cancer Res Clin Oncol 118: 269–275
van Valen F, Keck E, Jürgens H, Winkelmann W 1987 Beta-adrenergic agonist- and prostaglandin-mediated regulation of cAMP levels in Ewing's sarcoma cells in culture. Biochem Biophys Res Commun 146: 685–691
Whang-Peng J, Triche TJ, Knutsen T, Miser J, Douglass EC, Israel MA 1984 Chromosome translocation in peripheral neuroepithelioma. N Engl J Med 311: 584–585
Kodama K, Doi O, Higashiyama M, Yokouchi H, Tateishi R, Mori Y 1994 Differentiation of a Ewing's sarcoma cell line towards neural and mesenchymal cell lineages. Jpn J Cancer Res 85: 335–338
Baersch G, Möllers T, Hötte A, Dockhorn-Dworniczak B, Rübe C, Ritter J, Jürgens H, Vormoor J 1997 Good engraftment of B-cell precursor ALL in NOD-SCID mice. Klin Pädiatr 209: 178–185
Dockhorn-Dworniczak B, Schäfer KL, Dantcheva R, Blasius S, Winkelmann W, Burdach S, van-Valen F, Jürgens H, Böcker W 1994 Diagnostic value of the molecular genetic detection of the 11;22 translocation in Ewing's tumours. Virchows Arch 424: 107–112
Vormoor J, Lapidot T, Pflumio F, Risdon G, Patterson B, Broxmeyer HE, Dick JE 1994 Immature human cord blood progenitors engraft and proliferate to high levels in severe combined immunodeficient mice. Blood 83: 2489–2497
Vormoor J, Baersch G, Baumann M, Ritter J, Jürgens H 1998 Flow cytometric identification of candidate normal stem cell populations in CD45-negative B-cell precursor acute lymphoblastic leukaemia (ALL). Br J Haematol 100: 501–508
Taswell C 1981 Limiting dilution assay for the determination of immunocompetent cell frequencies. J Immunol 126: 1614–1619
Sanders RJ, Mainiero F, Giancotti FG 1998 The role of integrins in tumorigenesis and metastasis. Cancer Invest 16: 329–344
van der Loo JCM, Xiao X, McMillin D, Hashino K, Kato I, Williams DA 1998 VLA-5 is expressed by mouse and human long-term repopulating hematopoietic cells and mediates adhesion to extracellular matrix protein fibronectin. J Clin Invest 102: 1051–1061
Sell S, Pierce GB 1994 Maturation arrest of stem cell differentiation is a common pathway for the cellular origin of teratocarcinoma and epithelial cancers. Lab Invest 70: 6–22
van Hennik PB, de-Koning AE, Ploemacher RE 1999 Seeding efficiency of primitive human hematopoietic cells in nonobese diabetic/severe combined immune deficiency mice: Implications for stem cell frequency assessment. Blood 94: 3055–3061
Lapidot T, Grunberger T, Vormoor J, Estrov Z, Kizelstein P, Bunin N, Zaizov R, Williams D, Freedman MH 1996 Identification of human juvenile chronic myelogenous leukemia cells capable of initiating the disease in primary and secondary SCID mice. Blood 88: 2655–2664
Cavazzana AO, Miser JS, Jefferson J, Triche TJ 1987 Experimental evidence for a neural origin of Ewing's sarcoma of bone. Am J Pathol 127: 507–518
Dehner LP 1993 Primitive neuroectodermal tumor and Ewing's sarcoma. Am J Surg Pathol 17: 1–13
Jürgens H, Exener U, Gadner H, Harms D, Michaelis J, Sauer R, Treuner J, Voute T, Winkelmann W, Winkler K, Göbel U 1988 Multidisciplinary treatment of Ewing's sarcoma of bone. A 6-year experience of a European Cooperative Trial. Cancer 61: 23–32
Antoneli CBG, Costa CML, Camargo Bd, Sredni ST, Alfer W, Chojniak R 1998 Primitive neuroectodermal tumor (PNET)/Extraosseous Ewing sarcoma of the kidney. Med Ped Oncol 30: 303–307
Marley EF, Liapis H, Humphrey PA, Nadler RB, Siegel CL, Zhu X, Brandt JM, Dehner LP 1997 Primitive neuroectodermal tumor of the kidney—another enigma: a pathologic, immunohistochemical, and molecular diagnostic study. Am J Surg Pathol 21: 354–359
Sheaff M, McManus A, Scheimberg I, Paris A, Shipley J, Baithun S 1997 Primitive neuroectodermal tumor of the kidney confirmed by fluorescence in situ hybridization. Am J Surg Pathol 21: 461–468
Kawauchi S, Fukuda T, Miyamoto S, Yoshioka J, Shirahama S, Saito T, Tsukamoto N 1998 Peripheral primitive neuroectodermal tumor of the ovary confirmed by CD99 immunostaining, karyotypic analysis, and RT-PCR for EWS/FLI-1 chimeric mRNA. Am J Surg Pathol 22: 1417–1422
Young RH, Scully RE 1990 Sarcomas metastatic to the ovary: a report of 21 cases. Int J Gynecol Pathol 9: 231–252
Young RH, Kozakewich HP, Scully RE 1993 Metastatic ovarian tumors in children: a report of 14 cases and review of the literature. Int J Gynecol Pathol 12: 8–19
Acknowledgements
The authors thank Prof. J.E. Dick, Toronto, Ontario, Canada, Prof. A.L. Harris, O-ford, U.K., and Prof. J. Kienast and Dr. M. Paulussen, Münster, Germany, for stimulating discussions and for critically reviewing the manuscript, and Prof. K.-D. Richter, Central Animal Care Facility, Münster, Germany, for his continuing support. We are also grateful to our technicians Andrea Hötte, Frauke Schmidt, and Tanja Möllers for technical help.
Author information
Authors and Affiliations
Additional information
Supported by grants #10-0995-Vo 1 and #10-1252-Vo 2 from the Deutsche Krebshilfe.
Rights and permissions
About this article
Cite this article
Vormoor, J., Baersch, G., Decker, S. et al. Establishment of an In Vivo Model for Pediatric Ewing Tumors by Transplantation into NOD/ scid Mice. Pediatr Res 49, 332–341 (2001). https://doi.org/10.1203/00006450-200103000-00006
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1203/00006450-200103000-00006
This article is cited by
-
Ewing sarcoma dissemination and response to T-cell therapy in mice assessed by whole-body magnetic resonance imaging
British Journal of Cancer (2013)
-
The ganglioside antigen GD2 is surface-expressed in Ewing sarcoma and allows for MHC-independent immune targeting
British Journal of Cancer (2012)
-
Basic fibroblast growth factor in the bone microenvironment enhances cell motility and invasion of Ewing's sarcoma family of tumours by activating the FGFR1–PI3K–Rac1 pathway
British Journal of Cancer (2010)
-
Successful high-resolution animal positron emission tomography of human Ewing tumours and their metastases in a murine xenograft model
European Journal of Nuclear Medicine and Molecular Imaging (2006)
-
PI3K/AKT is involved in mediating survival signals that rescue Ewing tumour cells from fibroblast growth factor 2-induced cell death
British Journal of Cancer (2005)