Abstract
Background:
The major advantages of urine-based assays are their non-invasive character and ability to monitor prostate cancer (CaP) with heterogeneous foci. While the test for the prostate cancer antigen 3 (PCA3) is commercially available, the aim of our research was to test other putative urine markers in multiplex settings (AMACR (α-methylacyl-CoA racemase), EZH2 (enhancer of zeste homolog 2), GOLM1 (golgi membrane protein 1), MSMB (microseminoprotein, β), SPINK1 (serine peptidase inhibitor) and TRPM8 (transient receptor potential cation channel, subfamily M, member 8)).
Methods:
Expression of the candidate biomarkers was studied in sedimented urine using quantitative reverse transcriptase polymerase chain reaction in two sets of patients with and without restriction on serum PSA levels.
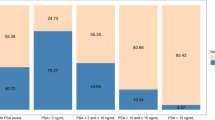
Results:
We confirmed that PCA3 is an independent predictor of cancer in the patients without restriction of serum PSA values (set 1, n=176, PSA=0.1–587 ng ml–1). However, AMACR was the only parameter that differentiated CaP from non-CaP patients with serum PSA between 3 and 15 ng ml–1 (set 2, n=104). The area under curve (AUC) for this gene was 0.645 with both sensitivity and specificity at 65%. Further improvement was achieved by multivariate logistic regression analysis, which identified novel duplex (TRPM8 and MSMB), triplex (plus AMACR) and quadriplex (plus PCA3) models for the detection of early CaPs (AUC=0.665, 0.726 and 0.741, respectively).
Conclusions:
Novel quadriplex test could be implemented as an adjunct to serum PSA or urine PCA3 and this could improve decision making for diagnostics in the case of ‘PSA dilemma’ patients.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 4 print issues and online access
$259.00 per year
only $64.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
van Gils MP, Stenman UH, Schalken JA, Schroder FH, Luider TM, Lilja H et al. Innovations in serum and urine markers in prostate cancer current European research in the P-Mark project. Eur Urol 2005; 48: 1031–1041.
Landers KA, Burger MJ, Tebay MA, Purdie DM, Scells B, Samaratunga H et al. Use of multiple biomarkers for a molecular diagnosis of prostate cancer. Int J Cancer 2005; 114: 950–956.
Schmidt U, Fuessel S, Koch R, Baretton GB, Lohse A, Tomasetti S et al. Quantitative multi-gene expression profiling of primary prostate cancer. Prostate 2006; 66: 1521–1534.
Laxman B, Tomlins SA, Mehra R, Morris DS, Wang L, Helgeson BE et al. Noninvasive detection of TMPRSS2:ERG fusion transcripts in the urine of men with prostate cancer. Neoplasia 2006; 8: 885–888.
Laxman B, Morris DS, Yu J, Siddiqui J, Cao J, Mehra R et al. A first-generation multiplex biomarker analysis of urine for the early detection of prostate cancer. Cancer Res 2008; 68: 645–649.
Schneider S, Voigt S, Fussel S, Lohse-Fischer A, Tomasetti S, Haase M et al. [Molecular genetic markers for prostate cancer. Evidence in fine needle biopsies for improved confirmation of the diagnosis]. Urologe A 2008; 47: 1208–1211.
Ouyang B, Bracken B, Burke B, Chung E, Liang J, Ho SM . A duplex quantitative polymerase chain reaction assay based on quantification of alpha-methylacyl-CoA racemase transcripts and prostate cancer antigen 3 in urine sediments improved diagnostic accuracy for prostate cancer. J Urol 2009; 181: 2508–2513; discussion 2513–2514.
Noutsias M, Rohde M, Block A, Klippert K, Lettau O, Blunert K et al. Preamplification techniques for real-time RT-PCR analyses of endomyocardial biopsies. BMC Mol Biol 2008; 9: 3.
Pfaffl MW . A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 2001; 29: e45.
Hoffman RM, Gilliland FD, Adams-Cameron M, Hunt WC, Key CR . Prostate-specific antigen testing accuracy in community practice. BMC Fam Pract 2002; 3: 19.
van Gils MP, Hessels D, van Hooij O, Jannink SA, Peelen WP, Hanssen SL et al. The time-resolved fluorescence-based PCA3 test on urinary sediments after digital rectal examination; a Dutch multicenter validation of the diagnostic performance. Clin Cancer Res 2007; 13: 939–943.
Marks LS, Fradet Y, Deras IL, Blase A, Mathis J, Aubin SM et al. PCA3 molecular urine assay for prostate cancer in men undergoing repeat biopsy. Urology 2007; 69: 532–535.
Haese A, de la Taille A, van Poppel H, Marberger M, Stenzl A, Mulders PF et al. Clinical utility of the PCA3 urine assay in European men scheduled for repeat biopsy. Eur Urol 2008; 54: 1081–1088.
Thompson IM, Ankerst DP . Prostate-specific antigen in the early detection of prostate cancer. CMAJ 2007; 176: 1853–1858.
Rogers CG, Yan G, Zha S, Gonzalgo ML, Isaacs WB, Luo J et al. Prostate cancer detection on urinalysis for alpha methylacyl coenzyme a racemase protein. J Urol 2004; 172: 1501–1503.
Prior C, Guillen-Grima F, Robles JE, Rosell D, Fernandez-Montero JM, Agirre X et al. Use of a combination of biomarkers in serum and urine to improve detection of prostate cancer. World J Urol 2010; 28: 681–686.
Tomlins SA, Rhodes DR, Yu J, Varambally S, Mehra R, Perner S et al. The role of SPINK1 in ETS rearrangement-negative prostate cancers. Cancer Cell 2008; 13: 519–528.
Varambally S, Laxman B, Mehra R, Cao Q, Dhanasekaran SM, Tomlins SA et al. Golgi protein GOLM1 is a tissue and urine biomarker of prostate cancer. Neoplasia 2008; 10: 1285–1294.
Zhang L, Barritt GJ . TRPM8 in prostate cancer cells: a potential diagnostic and prognostic marker with a secretory function? Endocr Relat Cancer 2006; 13: 27–38.
Bai VU, Murthy S, Chinnakannu K, Muhletaler F, Tejwani S, Barrack ER et al. Androgen regulated TRPM8 expression: a potential mRNA marker for metastatic prostate cancer detection in body fluids. Int J Oncol 2010; 36: 443–450.
Teni TR, Sheth AR, Kamath MR, Sheth NA . Serum and urinary prostatic inhibin-like peptide in benign prostatic hyperplasia and carcinoma of prostate. Cancer Lett 1988; 43: 9–14.
Tsurusaki T, Koji T, Sakai H, Kanetake H, Nakane PK, Saito Y . Cellular expression of beta-microseminoprotein (beta-MSP) mRNA and its protein in untreated prostate cancer. Prostate 1998; 35: 109–116.
Whitaker HC, Warren AY, Eeles R, Kote-Jarai Z, Neal DE . The potential value of microseminoprotein-beta as a prostate cancer biomarker and therapeutic target. Prostate 2010; 70: 333–340.
Kuefer R, Varambally S, Zhou M, Lucas PC, Loeffler M, Wolter H et al. alpha-Methylacyl-CoA racemase: expression levels of this novel cancer biomarker depend on tumor differentiation. Am J Pathol 2002; 161: 841–848.
Rubin MA, Bismar TA, Andren O, Mucci L, Kim R, Shen R et al. Decreased alpha-methylacyl CoA racemase expression in localized prostate cancer is associated with an increased rate of biochemical recurrence and cancer-specific death. Cancer Epidemiol Biomarkers Prev 2005; 14: 1424–1432.
Nguyen PN, Violette P, Chan S, Tanguay S, Kassouf W, Aprikian A et al. A panel of TMPRSS2:ERG fusion transcript markers for urine-based prostate cancer detection with high specificity and sensitivity. Eur Urol 2011; 59: 407–414.
Saramaki OR, Tammela TL, Martikainen PM, Vessella RL, Visakorpi T . The gene for polycomb group protein enhancer of zeste homolog 2 (EZH2) is amplified in late-stage prostate cancer. Genes Chromosomes Cancer 2006; 45: 639–645.
Varambally S, Dhanasekaran SM, Zhou M, Barrette TR, Kumar-Sinha C, Sanda MG et al. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature 2002; 419: 624–629.
Specht K, Richter T, Muller U, Walch A, Werner M, Hofler H . Quantitative gene expression analysis in microdissected archival formalin-fixed and paraffin-embedded tumor tissue. Am J Pathol 2001; 158: 419–429.
Acknowledgements
This work was supported by Grants NS 9940-4 from the Czech Ministry of Health and MSM 6198959216 from the Czech Ministry of Education and EU infrastructure support CZ.1.05/2.1.00/01.0030. Tamar Jamaspishvili was also supported by GACR 303/09/H048 from the Grant Agency of the Czech Republic and LF_2010_006. We sincerely thank Jana Holinkova for her excellent technical assistance.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on the Prostate Cancer and Prostatic Diseases website
Supplementary information
Rights and permissions
About this article
Cite this article
Jamaspishvili, T., Kral, M., Khomeriki, I. et al. Quadriplex model enhances urine-based detection of prostate cancer. Prostate Cancer Prostatic Dis 14, 354–360 (2011). https://doi.org/10.1038/pcan.2011.32
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/pcan.2011.32
Keywords
This article is cited by
-
Approaches to urinary detection of prostate cancer
Prostate Cancer and Prostatic Diseases (2019)
-
Using gene expression from urine sediment to diagnose prostate cancer: development of a new multiplex mRNA urine test and validation of current biomarkers
BMC Cancer (2016)
-
Linkage Analysis of Extended High-Risk Pedigrees Replicates a Cutaneous Malignant Melanoma Predisposition Locus on Chromosome 9q21
Journal of Investigative Dermatology (2013)