Abstract
Rhabdomyosarcoma, one of the most common childhood sarcomas, is comprised of two main subtypes, embryonal and alveolar (ARMS). ARMS, the more aggressive subtype, is primarily characterized by the t(2;13)(p35;p14) chromosomal translocation, which fuses two transcription factors, PAX3 and FOXO1 to generate the oncogenic fusion protein PAX3-FOXO1. Patients with PAX3-FOXO1-postitive tumors have a poor prognosis, in part due to the enhanced local invasive capacity of these cells, which leads to the increased metastatic potential for this tumor. Despite this knowledge, little is known about the role that the oncogenic fusion protein has in this increased invasive potential. In this report we use large-scale comparative transcriptomic analyses in physiologically relevant primary myoblasts to demonstrate that the presence of PAX3-FOXO1 is sufficient to alter the expression of 70 mRNA and 27 miRNA in a manner predicted to promote cellular invasion. In contrast the expression of PAX3 alters 60 mRNA and 23 miRNA in a manner predicted to inhibit invasion. We demonstrate that these alterations in mRNA and miRNA translate into changes in the invasive potential of primary myoblasts with PAX3-FOXO1 increasing invasion nearly 2-fold while PAX3 decreases invasion nearly 4-fold. Taken together, these results allow us to build off of previous reports and develop a more expansive molecular model by which the presence of PAX3-FOXO1 alters global gene regulatory networks to enhance the local invasiveness of cells. Further, the global nature of our observed changes highlights the fact that instead of focusing on a single-gene target, we must develop multi-faceted treatment regimens targeting multiple genes of a single oncogenic phenotype or multiple genes that target different oncogenic phenotypes for tumor progression.
Similar content being viewed by others
Introduction
Rhabdomyosarcoma (RMS), which accounts for nearly half of childhood soft tissue sarcomas, is comprised of two main subtypes: embryonal rhabdomyosarcoma and alveolar (ARMS), each defined by its unique histology, clinical presentation and prognosis.1 ARMS, the more aggressive subtype, is primarily defined by the t(2;13)(p35;p14) chromosomal translocation,2 which generates the oncogenic fusion protein PAX3-FOXO1.3, 4 PAX3-FOXO1 has altered molecular activities relative to wild-type PAX3, including being a more potent transcriptional activator,5 being unresponsive to normal PAX3 co-regulators6 and having greater post-translational stability upon the induction of myogenic differentiation.7 These aberrant molecular activities are believed to contribute to altered gene regulation, including the activation of genes not normally regulated by PAX38 and increased expression of other genes relative to PAX3,9, 10 which taken together is believed to contribute to ARMS tumor phenotypes.11
Patients diagnosed with PAX3-FOXO1-positive ARMS have a 4-year survival rate of 8%.12 This poor prognosis stems in part from these tumor cells having a higher incidence of localized invasion,12 which may then lead to heightened aggressiveness and an increased propensity for metastasis. The presence of PAX3-FOXO1 is known to enhance the invasive potential of cells,13 possibly through its ability to alter the expression of multifunctional genes that contribute, in part to invasion in other tumor types, including MET,10 FGFR4,14 IGF215 and IGFBP5.15 Despite these circumstantial correlations, to date only a single report demonstrates that the PAX3-FOXO1 altered expression of a gene, the cannabinoid receptor 1, directly contributes to the invasive capacity in ARMS.16 However, these results were derived from the expression of the oncogenic fusion protein in established tumor cell lines13 or in primary myoblasts that genetically contained compensatory oncogenic mutations.16 Further, these reports either did not examine altered gene expression13 or focused their study on changes in the expression of a single gene.16 While these reports are noteworthy and of importance, they provide little information to describe how the expression of PAX3-FOXO1 in the absence of any other compensatory mutations globally alters mRNA expression patterns to contribute to invasion. Further, to date no studies have directly examined how the presence of PAX3-FOXO1 affects microRNA (miRNA) expression and how these changes contribute to the invasive capacity of myoblasts.
In this study we utilized physiologically relevant wild-type primary myoblasts along with large-scale comparative transcriptomic analyses to examine how the expression of PAX3-FOXO1 or PAX3 alters global mRNA and miRNA expression profiles and how these changes contribute to the invasive potential of these cells. We report here that the expression of the oncogenic fusion protein is sufficient to alter the expression of 70 mRNA and 27 miRNA in such a way that would be expected to promote cellular invasion. In contrast, the expression of PAX3 elicits mRNA and miRNA expression changes that would be expected to inhibit cellular invasion. We found that these mRNA and miRNA changes translate into biological effects, with the expression of PAX3-FOXO1 enhancing and the expression of PAX3 inhibiting primary myoblast invasion. Taken together, these results provide a more expansive picture to describe the increased localized invasion seen with t(2;13)(q35;q14) positive ARMS tumors, and describes how the presence of PAX3-FOXO1 may contribute to higher levels of metastasis in these patients.
Results and Discussion
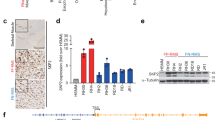
To understand how PAX3-FOXO1 affects global mRNA and miRNA expression, we stably transduced passage-matched wild-type mouse primary myoblasts with the MSCV-IRES-puromycin retroviral vector (negative control), or the same retroviral vector expressing FLAG epitope-tagged PAX3 (FLAG-PAX3) or FLAG-PAX3-FOXO1, a tag previously shown to not affect Pax3 or Pax3-FOXO1 function.6, 17 The puromycin selected cells were harvested from three independent transductions and pooled, resulting in a single mixed population for each individual construct, which removes the potential for variability that may occur from clonal effects. The level of PAX3-FOXO1 expression is equivalent to the level of expression of the fusion protein in ARMS tumor cell lines (Figure 1 and Dietz et al.18, 19) and is therefore directly relevant to the role of the oncogenic fusion protein in ARMS. This model allows us to use a physiologically relevant cell system in the absence of any complimentary transforming mutations to determine the specific effects of PAX3-FOXO1 on oncogenic phenotypes.
Protein expression (a) and quantitative RT–PCR analyses for (b) select mRNA and (c) select miRNA. Mouse primary myoblasts were isolated from 2- to 4-day-old C57/Bl6 mice as previously described.53 Cells were grown as previously described7, 17, 18, 19, 53 and were passage-matched to prevent possible differences due to passage conditions. Mouse primary myoblasts were stably transduced as previously described6, 53 with the MSCV-IRES-puromycin empty vector, vector containing FLAG epitope-tagged Pax3 (FLAG-Pax3) or FLAG-PAX3-FOXO1. Three days post transduction, cells were selected using puromycin, as previously described.19 The stably transduced cells were harvested and pooled from three independent transductions to create a single population that express each construct. (a) Total cell extracts made, as previously described.17, 18, 19, 53 Equal amounts of total cell lysates (12 μg) were separated by 8% SDS–PAGE and analyzed by western blot analysis using antibodies specific for Pax3,54 as previously described.18, 19 (b,c) Total RNA was isolated from the stably transduced proliferating primary myoblasts (empty vector (white bars), PAX3 (gray bars) or PAX3-FOXO1 (black bars)) using the miRNeasy mini kit (Qiagen, Madison, WI, USA), allowing for the isolation of RNA <30 bp in length, according to the manufacturer’s specifications. Equal amounts of total RNA (100 ng) were used for cDNA synthesis using the iScript cDNA synthesis kit (Bio-Rad, Hercules, CA, USA) for mRNA (b) or the Taqman miRNA reverse transcription kit (Applied Biosystems, Foster City, CA, USA) for miRNA. (c) The qRT–PCR was performed on the resulting cDNA using the CFX96 Touch Real-Time PCR Detection System (Bio-Rad) using commercially available primer/probe sets and the Applied Biosystems Universal Master Mix (Applied Biosystems), according to the manufacturer’s specifications. All results were normalized for GAPDH (mRNA) or the U6 small nuclear RNA (miRNA) and reported as fold expression relative to the results obtained for cells stably transduced with the empty vector. In all cases, analyses were performed comparing each sample with the empty vector control (*P=0.009, **P=0.001, ***P=0.0001).
We performed mRNA and miRNA deep-sequencing analyses on total RNA isolated from three independent growths of stably transduced cells and utilized the resulting data to perform comparative transcriptomic analyses to understand how each protein alters expression profiles to exert their effects on the invasive capacity of cells. For both the mRNA and miRNA analyses the data used for subsequent studies were limited to (1) those genes or miRNA displaying statistically significant differences (P<0.05, as determined by the Galaxy Cuffdiff program (mRNA) or miRNAKey (miRNA)), (2) transcripts whose expression was present in both data sets being analyzed to rule out potential artifactual differences resulting from depth of read and (3) transcripts or miRNA that exhibited at least 2-fold differences in expression either up or downregulated.
We found a total of 480 mRNA whose expression changed in a PAX3-FOXO1-dependent manner (276 downregulated and 204 upregulated) relative to the empty vector negative control (data not shown). We performed a PubMed search on each of the 480 mRNA, using the gene name and the search term ‘invasion’ to determine if they were experimentally proven to contribute to cellular invasion. We found that 70 of the 480 altered genes (14.5%) are involved in regulating the invasive capacity of cells (Table 1). Forty-three of these genes have literature evidence demonstrating their role in promoting cellular invasion, with these altered genes being split nearly equally between being upregulated (19/43; 44%) or downregulated (24/43; 56%) in a PAX3-FOXO1-dependent manner. In a similar manner, 27 genes have literature evidence to support their role in inhibiting cellular invasion, with 21 of these genes (nearly 80%) being downregulated in a PAX3-FOXO1-dependent manner. Finally, 17 of the 70 differentially expressed genes (nearly 25%) contain PAX3-FOXO1 binding sites in their proximal promoters, as previously described20 (Table 1, c), four of these genes were previously demonstrated to be regulated by PAX3-FOXO1, including cannabinoid receptor 1,16 FGFR4,20 IGF221 and IGFBP515 (Table 1, b), and 21 of the 70 (30%) genes have altered gene expression levels consistent with changes seen in human tumor samples22, 23, 24, 25 (Table 1, a).
An initial examination of the distribution of mRNA whose levels are altered upon the expression of the fusion protein would suggest that PAX3-FOXO1 would primarily exert its invasive effect13 by decreasing the expression of genes important for inhibiting invasion. However, it is interesting to note that although only 44% of the genes that promote invasion are upregulated, nearly half of these 19 upregulated genes (8/19–42%) are increased >6-fold, including the previously reported cannabinoid receptor16 (cannabinoid receptor 1–6.92-fold), with the top four genes being upregulated >20-fold. Therefore, this data suggest that PAX3-FOXO1 exerts its effects on invasive capacity by not only decreasing the expression of a large number of inhibitory genes, but by simultaneously greatly increasing the expression of key genes that promote invasion, including genes that encode for proteins involved in cytoskeletal organization (CAP6–33.45-fold), cadherins (CDH6–23.23-fold), extracellular matrix metalloproteases (ADAMTS1–21.98-fold) and cell adhesion proteins (MSLN – 20.93-fold).
A similar analysis found 399 mRNA change in a PAX3-dependent manner (276 downregulated and 123 upregulated) relative to the empty vector negative control (data not shown). A similar PubMed search revealed that 60 of the 399 genes (15%) are involved with regulating invasion (Table 1). Thirty-eight of these genes have a demonstrated role in promoting invasion, with a majority of these genes (25/38; 66%) being downregulated. Further, 22 mRNA were demonstrated to inhibit invasion, with 6 of these genes being upregulated and 16 being downregulated. Finally, four of the differentially expressed genes were demonstrated in the literature to be directly regulated by PAX3, including Ahr,26 IGF1R,20 EPHA227 and MET28 (Table 1, b). Although a smaller number of these inhibitory genes are upregulated, one of them is upregulated >15-fold (metallopeptidase Mme—15.17-fold). In contrast to the results seen with PAX3-FOXO1, these data suggest that PAX3 would be expected to inhibit invasive capacity, primarily through the downregulation of genes that promote this biological event.
A comparative transcriptomic analysis of the miRNA data identified a total of 84 miRNAs whose expression changed in a PAX3-FOXO1-dependent manner (46 downregulated and 38 upregulated) relative to the empty vector negative control (data not shown). A PubMed search of each of the individual 84 miRNA, using the miRNA name and the search term ‘invasion’, indicated that 10 of these miRNA promote cellular invasion (Table 2). Of these miRNA, 9/10 (90%) have an increased PAX3-FOXO1-dependent expression with the top two being increased >20-fold. In a similar manner, 17 miRNA are important for inhibiting cellular invasion, of which 16/17 (94%) are downregulated, with the top four being downregulated >12-fold. In conjunction with the results of our PubMed search, which also described the target genes responsible for the invasive effect of the miRNA, we used miRTarBase29 to identify known target genes whose biological function may contribute to an invasive phenotype, with validation on miRTarBase by at least two independent experimental methods. Interestingly, only a small number of the altered miRNA have the expected inverse correlation to our observed changes in mRNA expression (miR-222/miR-221 and TIMP2, and miR-362 and CD82).
A similar analysis determined a total of 58 miRNA whose expression changed upon the expression of PAX3 (25 downregulated and 33 upregulated) relative to the empty vector negative control (data not shown). Of these genes, a PubMed search determined that 7 are important for promoting while 16 inhibit cellular invasion. Of those miRNA, 5/7 (71%) that promote invasion are decreased whereas 10/16 (63%) that inhibit invasion are increased, with the top inhibitory miRNA being increased >15-fold. Finally, for both PAX3-FOXO1- and PAX3-dependent miRNA changes, the literature provides direct evidence for the genes they target in order to exert their effects on invasion (Table 2). As seen with PAX3-FOXO1 changes, only one of the PAX3-altered miRNA has the expected inverse correlation to mRNA expression (miR-206 and MET, Tables 1 and 2). Interestingly, three sets of miRNA are present as clusters in the mouse genome and have similar changes in expression. These include miRNA 222 and 221, which are upregulated to a similar extent by PAX3-FOXO1 while being downregulated to a similar extent by PAX3 (Table 2, a); miRNA 362 and 532, which are downregulated to a similar extent by PAX3-FOXO1 but are unaffected by PAX3 (Table 2, b); and miRNA 133b and 206, which are unaffected by PAX3-FOXO1 are upregulated to a similar extent by PAX3 (Table 2, c).
To validate the observed changes, we performed a quantitative RT–PCR analysis on a subset of mRNA and miRNA. We tested IGF2, which was reported to promote cellular invasion in a variety of tumors,30, 31, 32, 33 and CDK1, which has alternative roles in inhibiting cellular invasion in different tumor models.34, 35, 36, 37, 38 We also examined two miRNA (miR-196a-5p and miR301a-3p), both demonstrated to promote cellular invasion.39, 40, 41, 42, 43, 44 We observed quantitative and significant changes in expression for all of the mRNA and miRNA examined that are consistent with our mRNA deep-sequencing results (Figure 1).
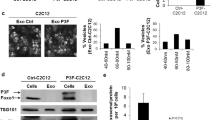
Our comparative transcriptomic results suggest that the mRNA and miRNA changes induced by the oncogenic fusion protein would be predicted to promote cellular invasion, whereas those changes that occur in a PAX3-dependent manner should inhibit the invasive capacity of cells. Therefore, we used a standard invasion assay to determine whether our observed mRNA and miRNA changes translate into experimental differences in the invasive potential of primary myoblasts. Consistent with our mRNA and miRNA changes, we observed a nearly 2-fold increase in primary myoblasts expressing PAX3-FOXO1, consistent with the previous reports,16 whereas cells expressing PAX3 had a nearly 4-fold decrease in invasive potential (Figure 2).
Pax3-FOXO1 promotes whereas Pax3 inhibits primary myoblast invasive capacity. Invasive capacity was determined using stably transduced proliferating primary myoblasts (empty vector (white bars), PAX3 (gray bars) or PAX3-FOXO1 (black bars)) using the BD Biocoat Tumor Invasion System (Becton Dickinson, Franklin Lakes, NJ, USA). About 50 000 cells suspended in proliferation media were added to the insert plate with proliferation media supplemented with hepatocyte growth factor (hHGF, PeproTech, Rocky Hill, NJ, USA) at 25 ng/ml being used as the chemoattractant. After 24 h of incubation, the insert system was transferred to a second 24-well plate containing calcein AM in Hank's balanced salt solution (HBSS) that enabled the fluorescent labeling of cells that invaded through the Matrigel matrix. Fluorescence of the invaded cells was read at wavelengths of 494/517 (Ex/Em) using a Synergy HT multi-well microplate reader (BioTek, Winooski, VT, USA). P-values were computed using non-parametric one-way ANOVA analysis comparing all samples with results obtained, with cells expressing empty vector (*P=0.03, **P=0.001). ANOVA, analysis of variance.
Taken together, our results build off of previous work, in which a single gene was examined,16 and show that the sole expression of PAX3-FOXO1 in the absence of any complimentary genetic mutations is capable of globally altering mRNA and miRNA levels to promote the invasive capacity of primary myoblasts. Further, this is the first study to examine how the oncogenic fusion protein alters miRNA levels, which combined with our global mRNA results allow us to develop a more expansive picture of the underlying regulatory mechanisms by which the expression of PAX3-FOXO1 promotes invasion.
In this regulatory mechanism, the somatic and random acquisition of the chromosomal translocation creates the fusion protein, which alters, either directly or indirectly, the expression of mRNA important for enhancing cellular invasion. PAX3-FOXO1 achieves this by both decreasing the expression of genes that inhibit invasion (80% of downregulated genes) while also greatly increasing the expression of nearly half of the altered genes that promote invasion (Table 1). However, our results demonstrate that miRNAs, which post-transcriptionally ‘fine tune’ gene expression, also have a significant role, as nearly all of the increased miRNAs promote invasion and a majority of the decreased miRNAs inhibit invasion. Further, a closer inspection of the data reveals that although some of the miRNA changes have an inverse correlation to target genes present in our results, there are only three miRNA (miR-221, miR-222 and miR-362) that have such a correlation with their target genes (TIMP2 and CD82, respectively). Therefore, instead of post-transcriptionally contributing to our observed mRNA changes, the altered miRNA target a different set of genes important for invasive capacity, thereby greatly increasing the number of affected genes.
The global nature of the mRNA and miRNA expression changes that result upon the sole expression of PAX3-FOXO1 provide a basis for how it may be necessary to rethink approaches to the development of therapies for ARMS. At present, many developmental ARMS therapies focus on attacking a single gene or pathway mechanistically located downstream of the fusion protein. However, given that the expression of PAX3-FOXO1 alters the expression of 70 different genes and 27 different miRNAs to affect the invasive potential of cells, it is not too surprising that such focused and targeted therapies are not proving effective in Phase I or Phase II clinical trials for ARMS.45, 46, 47, 48 It is conceivable that the loss of a single gene through these targeted and focused therapies could easily be compensated through the changes in nearly 100 other affected genes, thereby negating the effects of the treatment.
Work in the past few years identified multiple aspects of the invasive process as potential targets for therapy development. Along these lines we propose developing a multi-faceted regimen that targets several of these processes, targets that include tumor-promoting genes we found to be the most highly upregulated in our study (Table 1). These processes include cytoskeletal remodeling, which is mediated in part by the intracellular signaling cysteine proteases calpains49 (Capn6 is upregulated 33-fold in our study), cellular adhesion mediated by such molecules as mesothelin50 (Msln is upregulated 21-fold in our study) and matrix metalloproteases,51 in particular the Adamts family of proteases52 (Adamts1 is upregulated 22-fold and Adamts5 is upregulated 7.5-fold in our study). A regimen that minimally targets these three processes would inhibit the necessary biological events required for invasion and metastasis. Alternatively, inhibiting one of these events (for example, matrix metalloproteases) could serve as one arm of a novel multi-faceted regimen for the treatment of ARMS, a regiment that also targets other ARMS molecular processes such as inhibiting phosphorylation of PAX3-FOXO1,17 attacking aneuploid cells and preventing enhanced proliferation.
References
Horn RC Jr., Enterline HT . Rhabdomyosarcoma: a clinicopathological study and classification of 39 cases. Cancer 1958; 11: 181–199.
Punyko JA, Mertens AC, Baker KS, Ness KK, Robison LL, Gurney JG . Long-term survival probabilities for childhood rhabdomyosarcoma. A population-based evaluation. Cancer 2005; 103: 1475–1483.
Galili N, Davis RJ, Fredericks WJ, Mukhopadhyay S, Rauscher FJ 3rd, Emanuel BS et al. Fusion of a fork head domain gene to PAX3 in the solid tumour alveolar rhabdomyosarcoma. Nat Genet 1993; 5: 230–235.
Shapiro DN, Sublett JE, Li B, Downing JR, Naeve CW . Fusion of PAX3 to a member of the forkhead family of transcription factors in human alveolar rhabdomyosarcoma. Cancer Res 1993; 53: 5108–5112.
Fredericks WJ, Galili N, Mukhopadhyay S, Rovera G, Bennicelli J, Barr FG et al. The PAX3-FKHR fusion protein created by the t(2;13) translocation in alveolar rhabdomyosarcomas is a more potent transcriptional activator than PAX3. Mol Cell Biol 1995; 15: 1522–1535.
Hollenbach AD, Sublett JE, McPherson CJ, Grosveld G . The Pax3-FKHR oncoprotein is unresponsive to the Pax3-associated repressor hDaxx. Embo J 1999; 18: 3702–3711.
Miller PJ, Hollenbach AD . The oncogenic fusion protein Pax3-FKHR has a greater post-translational stability relative to Pax3 during early myogenesis. Biochim Biophys Acta 2007; 1770: 1450–1458.
Epstein JA, Song B, Lakkis M, Wang C . Tumor-specific PAX3-FKHR transcription factor, but not PAX3, activates the platelet-derived growth factor alpha receptor. Mol Cell Biol 1998; 18: 4118–4130.
Ayalon D, Glaser T, Werner H . Transcriptional regulation of IGF-I receptor gene expression by the PAX3-FKHR oncoprotein. Growth Horm IGF Res 2001; 11: 289–297.
Ginsberg JP, Davis RJ, Bennicelli JL, Nauta LE, Barr FG . Up-regulation of MET but not neural cell adhesion molecule expression by the PAX3-FKHR fusion protein in alveolar rhabdomyosarcoma. Cancer Res 1998; 58: 3542–3546.
Linardic CM . PAX3-FOXO1 fusion gene in rhabdomyosarcoma. Cancer Lett 2008; 270: 10–18.
Sorensen PH, Lynch JC, Qualman SJ, Tirabosco R, Lim JF, Maurer HM et al. PAX3-FKHR and PAX7-FKHR gene fusions are prognostic indicators in alveolar rhabdomyosarcoma: a report from the children's oncology group. J Clin Oncol 2002; 20: 2672–2679.
Anderson J, Ramsay A, Gould S, Pritchard-Jones K . PAX3-FKHR induces morphological change and enhances cellular proliferation and invasion in rhabdomyosarcoma. Am J Pathol 2001; 159: 1089–1096.
Lagha M, Kormish JD, Rocancourt D, Manceau M, Epstein JA, Zaret KS et al. Pax3 regulation of FGF signaling affects the progression of embryonic progenitor cells into the myogenic program. Genes Dev 2008; 22: 1828–1837.
Khan J, Bittner ML, Saal LH, Teichmann U, Azorsa DO, Gooden GC et al. cDNA microarrays detect activation of a myogenic transcription program by the PAX3-FKHR fusion oncogene. Proc Natl Acad Sci USA 1999; 96: 13264–13269.
Marshall AD, Lagutina I, Grosveld GC . PAX3-FOXO1 induces cannabinoid receptor 1 to enhance cell invasion and metastasis. Cancer Res 2011; 71: 7471–7480.
Loupe JM, Miller PJ, Ruffin DR, Stark MW, Hollenbach AD . Inhibiting phosphorylation of the oncogenic PAX3-FOXO1 reduces alveolar rhabdomyosarcoma phenotypes identifying novel therapy options. Oncogenesis 2015; 4: e145.
Dietz KN, Miller PJ, Hollenbach AD . Phosphorylation of serine 205 by the protein kinase CK2 persists on Pax3-FOXO1, but not Pax3, throughout early myogenic differentiation. Biochemistry 2009; 48: 11786–11795.
Dietz KN, Miller PJ, Iyengar AS, Loupe JM, Hollenbach AD . Identification of serines 201 and 209 as sites of Pax3 phosphorylation and the altered phosphorylation status of Pax3-FOXO1 during early myogenic differentiation. Int J Biochem Cell Biol 2011; 43: 936–945.
Cao L, Yu Y, Bilke S, Walker RL, Mayeenuddin LH, Azorsa DO et al. Genome-wide identification of PAX3-FKHR binding sites in rhabdomyosarcoma reveals candidate target genes important for development and cancer. Cancer Res 2010; 70: 6497–6508.
Khan J, Simon R, Bittner M, Chen Y, Leighton SB, Pohida T et al. Gene expression profiling of alveolar rhabdomyosarcoma with cDNA microarrays. Cancer Res 1998; 58: 5009–5013.
Davicioni E, Finckenstein FG, Shahbazian V, Buckley JD, Triche TJ, Anderson MJ . Identification of a PAX-FKHR gene expression signature that defines molecular classes and determines the prognosis of alveolar rhabdomyosarcomas. Cancer Res 2006; 66: 6936–6946.
De Pitta C, Tombolan L, Albiero G, Sartori F, Romualdi C, Jurman G et al. Gene expression profiling identifies potential relevant genes in alveolar rhabdomyosarcoma pathogenesis and discriminates PAX3-FKHR positive and negative tumors. Int J Cancer 2006; 118: 2772–2781.
Ebauer M, Wachtel M, Niggli FK, Schafer BW . Comparative expression profiling identifies an in vivo target gene signature with TFAP2B as a mediator of the survival function of PAX3/FKHR. Oncogene 2007; 26: 7267–7281.
Lae M, Ahn EH, Mercado GE, Chuai S, Edgar M, Pawel BR et al. Global gene expression profiling of PAX-FKHR fusion-positive alveolar and PAX-FKHR fusion-negative embryonal rhabdomyosarcomas. J Pathol 2007; 212: 143–151.
Zalc A, Rattenbach R, Aurade F, Cadot B, Relaix F . Pax3 and Pax7 play essential safeguard functions against environmental stress-induced birth defects. Dev Cell 2015; 33: 56–66.
Begum S, Emami N, Cheung A, Wilkins O, Der S, Hamel PA . Cell-type-specific regulation of distinct sets of gene targets by Pax3 and Pax3/FKHR. Oncogene 2005; 24: 1860–1872.
Epstein JA, Shapiro DN, Cheng J, Lam PY, Maas RL . Pax3 modulates expression of the c-Met receptor during limb muscle development. Proc Natl Acad Sci USA 1996; 93: 4213–4218.
Hsu SD, Tseng YT, Shrestha S, Lin YL, Khaleel A, Chou CH et al. miRTarBase update 2014: an information resource for experimentally validated miRNA-target interactions. Nucleic Acids Res 2014; 42: D78–D85.
Chen YW, Boyartchuk V, Lewis BC . Differential roles of insulin-like growth factor receptor- and insulin receptor-mediated signaling in the phenotypes of hepatocellular carcinoma cells. Neoplasia 2009; 11: 835–845.
Mu Q, Wang L, Yu F, Gao H, Lei T, Li P et al. Imp2 regulates GBM progression by activating IGF2/PI3K/Akt pathway. Cancer Biol Ther 2015; 16: 623–633.
Pivonello C, Negri M, De Martino MC, Napolitano M, de Angelis C, Provvisiero DP et al. The dual targeting of insulin and insulin-like growth factor 1 receptor enhances the mTOR inhibitor-mediated antitumor efficacy in hepatocellular carcinoma. Oncotarget 2016; 7: 9718–9731.
Sciacca L, Mineo R, Pandini G, Murabito A, Vigneri R, Belfiore A . In IGF-I receptor-deficient leiomyosarcoma cells autocrine IGF-II induces cell invasion and protection from apoptosis via the insulin receptor isoform A. Oncogene 2002; 21: 8240–8250.
Chang WL, Yu CC, Chen CS, Guh JH . Tubulin-binding agents down-regulate matrix metalloproteinase-2 and -9 in human hormone-refractory prostate cancer cells - a critical role of Cdk1 in mitotic entry. Biochem Pharmacol 2015; 94: 12–21.
Vanan I, Dong Z, Tosti E, Warshaw G, Symons M, Ruggieri R . Role of a DNA damage checkpoint pathway in ionizing radiation-induced glioblastoma cell migration and invasion. Cell Mol Neurobiol 2012; 32: 1199–1208.
Wei Y, Chen YH, Li LY, Lang J, Yeh SP, Shi B et al. CDK1-dependent phosphorylation of EZH2 suppresses methylation of H3K27 and promotes osteogenic differentiation of human mesenchymal stem cells. Nat Cell Biol 2011; 13: 87–94.
Zhang C, Elkahloun AG, Robertson M, Gills JJ, Tsurutani J, Shih JH et al. Loss of cytoplasmic CDK1 predicts poor survival in human lung cancer and confers chemotherapeutic resistance. PLoS ONE 2011; 6: e23849.
Zhang L, Chen X, Stauffer S, Yang S, Chen Y, Dong J . CDK1 phosphorylation of TAZ in mitosis inhibits its oncogenic activity. Oncotarget 2015; 6: 31399–31412.
Egawa H, Jingushi K, Hirono T, Ueda Y, Kitae K, Nakata W et al. The miR-130 family promotes cell migration and invasion in bladder cancer through FAK and Akt phosphorylation by regulating PTEN. Sci Rep 2016; 6: 20574.
Fang Y, Sun B, Xiang J, Chen Z . MiR-301a promotes colorectal cancer cell growth and invasion by directly targeting SOCS6. Cell Physiol Biochem 2015; 35: 227–236.
Liu M, Du Y, Gao J, Liu J, Kong X, Gong Y et al. Aberrant expression miR-196a is associated with abnormal apoptosis, invasion, and proliferation of pancreatic cancer cells. Pancreas 2013; 42: 1169–1181.
Liu XH, Lu KH, Wang KM, Sun M, Zhang EB, Yang JS et al. MicroRNA-196a promotes non-small cell lung cancer cell proliferation and invasion through targeting HOXA5. BMC Cancer 2012; 12: 348.
Wang M, Li C, Yu B, Su L, Li J, Ju J et al. Overexpressed miR-301a promotes cell proliferation and invasion by targeting RUNX3 in gastric cancer. J Gastroenterol 2013; 48: 1023–1033.
Zhang W, Zhang T, Jin R, Zhao H, Hu J, Feng B et al. MicroRNA-301a promotes migration and invasion by targeting TGFBR2 in human colorectal cancer. J Exp Clin Cancer Res 2014; 33: 113.
Bagatell R, Norris R, Ingle AM, Ahern C, Voss S, Fox E et al. Phase 1 trial of temsirolimus in combination with irinotecan and temozolomide in children, adolescents and young adults with relapsed or refractory solid tumors: a Children's Oncology Group Study. Pediatr Blood Cancer 2014; 61: 833–839.
Geoerger B, Kieran MW, Grupp S, Perek D, Clancy J, Krygowski M et al. Phase II trial of temsirolimus in children with high-grade glioma, neuroblastoma and rhabdomyosarcoma. Eur J Cancer 2012; 48: 253–262.
Pappo AS, Vassal G, Crowley JJ, Bolejack V, Hogendoorn PC, Chugh R et al. A phase 2 trial of R1507, a monoclonal antibody to the insulin-like growth factor-1 receptor (IGF-1R), in patients with recurrent or refractory rhabdomyosarcoma, osteosarcoma, synovial sarcoma, and other soft tissue sarcomas: results of a Sarcoma Alliance for Research through Collaboration Study. Cancer 2014; 120: 2448–2456.
Weigel B, Malempati S, Reid JM, Voss SD, Cho SY, Chen HX et al. Phase 2 trial of cixutumumab in children, adolescents, and young adults with refractory solid tumors: a report from the Children's Oncology Group. Pediatr Blood Cancer 2014; 61: 452–456.
Leloup L, Wells A . Calpains as potential anti-cancer targets. Expert Opin Ther Targets 2011; 15: 309–323.
Pastan I, Hassan R . Discovery of mesothelin and exploiting it as a target for immunotherapy. Cancer Res 2014; 74: 2907–2912.
Cathcart J, Pulkoski-Gross A, Cao J . Targeting matrix metalloproteinases in cancer: bringing new life to old ideas. Genes Dis 2015; 2: 26–34.
Viapiano MS, Hockfield S, Matthews RT . BEHAB/brevican requires ADAMTS-mediated proteolytic cleavage to promote glioma invasion. J Neurooncol 2008; 88: 261–272.
Miller PJ, Dietz KN, Hollenbach AD . Identification of serine 205 as a site of phosphorylation on Pax3 in proliferating but not differentiating primary myoblasts. Protein Sci 2008; 17: 1979–1986.
Lam PY, Sublett JE, Hollenbach AD, Roussel MF . The oncogenic potential of the Pax3-FKHR fusion protein requires the Pax3 homeodomain recognition helix but not the Pax3 paired-box DNA binding domain. Mol Cell Biol 1999; 19: 594–601.
Acknowledgements
Funding for this project is from the National Cancer Institute grant R01CA138656, the Louisiana State University School of Medicine Research Enhancement Bridge Grant and the Louisiana Cancer Research Consortium (LCRC) (ADH). JZ has been partially supported by grants from the National Institute of General Medicine Sciences (NIGMS) grants P20GM103501, subproject #2, P30GM114732 and U54GM104940-01, and the National Institute on Minority Health and Health Disparities (NIMHD) grants P20MD004817 and U54MD006176-01. All deep sequencing was performed in the LCRC Translational Genomics Core facility.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Rights and permissions
Oncogenesis is an open-access journal published by Nature Publishing Group. This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Loupe, J., Miller, P., Bonner, B. et al. Comparative transcriptomic analysis reveals the oncogenic fusion protein PAX3-FOXO1 globally alters mRNA and miRNA to enhance myoblast invasion. Oncogenesis 5, e246 (2016). https://doi.org/10.1038/oncsis.2016.53
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/oncsis.2016.53
This article is cited by
-
Signaling pathways in Rhabdomyosarcoma invasion and metastasis
Cancer and Metastasis Reviews (2020)