Abstract
Drosophila myelodysplasia/myeloid leukemia factor (dMLF), a homolog of human MLF1, oncogene was first identified by yeast two-hybrid screen using the DNA replication-related element-binding factor (DREF) as bait. DREF is a transcription factor that regulates proliferation-related genes in Drosophila. It is known that overexpression of dMLF in the wing imaginal discs through the engrailed-GAL4 driver causes an atrophied wing phenotype associated with the induction of apoptosis. However, the precise mechanisms involved have yet to be clarified. Here, we found the atrophied phenotype to be suppressed by loss-of-function mutation of Drosophila Jun N-terminal kinase (JNK), basket (bsk). Overexpression of dMLF induced ectopic JNK activation in the wing disc monitored with the puckered-lacZ reporter line, resulting in induction of apoptosis. The DREF-binding consensus DRE sequence could be shown to exist in the bsk promoter. Chromatin immunoprecipitation assays in S2 cells with anti-dMLF IgG and quantitative real-time PCR revealed that dMLF binds specifically to the bsk promoter region containing the DRE sequence. Furthermore, using a transient luciferase expression assay, we provide evidence that knockdown of dMLF reduced bsk gene promoter activity in S2 cells. Finally, we show that dMLF interacts with DREF in vivo. Altogether, these data indicate that dMLF acts with DREF to stimulate the bsk promoter and consequently activates the JNK pathway to promote apoptosis.
Similar content being viewed by others
Introduction
Chromosomal translocations are frequently found in various human leukemias, and the aberrant proteins generated presumably interfere with the functions of their normal counterparts, in particular those associated with cellular growth and differentiation. Human myelodysplasia/myeloid leukemia factor 1 (hMLF1) was originally identified in the form of a fusion protein with nucleophosmin (NPM) generated by the t(3;5)(q25.1;q34) chromosomal translocation in the myelodysplastic syndrome and acute myeloid leukemia.1 NPM is a major nucleolar phosphoprotein that is significantly more abundant in tumors and proliferating cells than in normal resting cells.2, 3 It has also been reported that NPM shuttles between the nucleus and cytoplasm.4
The hMLF1 protein is mainly located in the cytoplasm, whereas its fused form with NPM is mostly located in the nucleus with highest levels in the nucleolus.1 The hMLF1 contains no recognizable domains or motifs except for a characteristic RSXSXP motif, the binding sequence for 14-3-3 protein,5, 6 that is involved in regulating cell division, apoptosis and differentiation. It has been reported that 14-3-3ζ interacts with hMLF1.7 In addition, it has been reported that hMLF1 associates with Madm, MLFIP/KLIP1, Manp and CSN3, a component of the COP9 signalosome, and regulates the cell cycle via the CSN/COP3 pathway.7, 8, 9, 10 However, the biochemical activity of hMLF1 has yet to be fully characterized.
The MLF protein is conserved among various species in animals.11, 12 In contrast to mammals that have two closely related proteins, hMLF1 and hMLF2,11 Drosophila has a single gene, dMLF, encoding a protein homologous to both hMLF1 and hMLF2. Drosophila myelodysplasia/myeloid leukemia factor (dMLF) was first identified by yeast two-hybrid screening using the DNA replication-related element-binding factor (DREF) as a bait.13 DREF is a transcription factor in Drosophila that regulates proliferation-related genes such as PCNA, Cyclin A, DNA polymerase α and others.14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25 The dMLF consists of 309 amino acid residues, and especially its central region (amino acids 96 to 202) displays high homology to hMLF1 (54% identity), hMLF2 (63%) and a mouse homolog, Hemopoietic lineage switch (HLS7) (59%).13 Furthermore, it is reported that this highly conserved region of dMLF is necessary for binding to DREF.13 The dMLF protein has also been reported to interact physically and genetically with Su(fu), a negative regulator of the Gli/Ci transcription factor involved in Hedgehog (Hh) signaling.26 In addition, it has also been reported that dMLF interacts with the Hh pathway component Cos2,26 and suppresses the rough eye phenotype induced by overexpression of DREF.13 Ectopic expression of dMLF in the developing eye imaginal disc using an eyeless-GAL4 driver resulted in a small eye phenotype rescued by coexpression of cyclin E, suggesting involvement of dMLF in cell-cycle regulation.27 Overexpression of dMLF in eye imaginal discs using a GMR-GAL4 driver also caused a rough eye phenotype in adults, and overexpression in wing imaginal discs induced programmed cell death and promoted transition through the S phase.26 It is therefore conceivable that dMLF plays multiple roles in cell growth, survival, apoptosis and gene transcription.
Other roles have been identified for dMLF. Thus, using a Drosophila model of polyglutamine disorders, it has been reported that overexpression of dMLF suppresses toxicity associated with an abnormally long polyglutamine tract expressed in the eye and central nervous system.28 dMLF reduced the recruitment of the CRE binding protein and Hsp70 into polyglutamine inclusions, both of these being among essential proteins apparently trapped in the inclusions.29 More recently, it has been shown that dMLF controls homeostasis of the Drosophila hematopoietic system by regulating the activity of the RUNX transcription factor Lozenge during development of crystal cells.30
In the present study, we found that dMLF is involved in the regulation of the Jun N-terminal kinase (JNK) pathway during Drosophila development. The JNK cascade is an intracellular signaling pathway in which the stress-activated kinases JNK kinase and JNK play essential roles.31 JNK signaling is involved in processes including cell proliferation and apoptosis.32, 33, 34 Apoptosis induced by JNK has an important role in the morphogenesis of the wing imaginal disc.35 JNK signaling controls the expression of target genes as those encoding the proapoptotic protein Reaper and the dual-specificity phosphatase Puckered (puc). In Drosophila, JNK kinase and JNK homologs are encoded by the genes hemipterous (hep) and basket (bsk), respectively.36, 37, 38 We have reported that the DREF-binding consensus, the DRE sequence, 5′-TATCGATA-3′, exists in the bsk promoter region and that DREF is required for bsk gene expression.25
In the present study, we further examined the roles of dMLF in the regulation of the JNK signaling pathway and demonstrated that dMLF acts with DREF in the bsk promoter to stimulate bsk expression and consequently activate the JNK pathway and apoptosis.
Results
dMLF genetically interacts with bsk
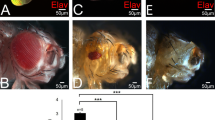
Transgenic fly lines overexpressing dMLF in the wing imaginal discs exhibit an atrophied phenotype of the posterior compartment of adult wings associated with caspase-3 activation and apoptosis in the wing imaginal discs.26 In the wing disc, JNK activation is usually linked to the activation of caspase-3.39 Therefore, we tested whether dMLF apoptotic effects in the wing discs could be due to the activation of the JNK pathway. Overexpression of dMLF in the wing discs using the posterior engrailed (en)-GAL4 driver (en-GAL4, UAS-dMLF flies) exhibited a severe reduction of the posterior part of the adult wing but not of the anterior compartment used here as a control (Figure 1b). Strikingly, half-dose reduction of bsk (en-GAL4, UAS-dMLF/bsk1) resulted in a suppression of this atrophied phenotype (Figure 1c). The same result was also seen with another allele of bsk, bsk2 (Figure 1d). These data suggest that dMLF apoptotic effect requires Bsk activity.
The bsk mutant suppresses the wing phenotype induced by overexpression of dMLF. (a) en-GAL4/+. Normal wings have five wing veins (L1, L2, L3, L4 and L5). The wing anterior includes L1, L2 and L3, whereas the wing posterior includes L4 and L5. (b) en-GAL4, UAS-dMLF/+. Overexpression of dMLF in wing imaginal discs results in an atrophied phenotype of the wing posterior. (c) en-GAL4, UAS-dMLF/bsk1. (d) en-GAL4, UAS-dMLF/bsk2. The aberrant wing phenotype was suppressed by crossing with loss-of -function alleles of bsk. The results suggest that dMLF positively regulates the JNK pathway.
Overexpression of dMLF induces Bsk (JNK) activation and apoptosis
The dMLF overexpression may act through Bsk to positively regulate the JNK pathway in the wing disc. Therefore, we monitored the effect of dMLF overexpression on the expression of puc, a gene highly expressed in Bsk-activated cells.39, 40 For that purpose, we used an enhancer trap line, pucE69, in which LacZ is inserted into the puc gene intron39, 40 so that puc enhancer activity can be monitored with reference to LacZ expression. It is well known that the puc gene is highly expressed in Bsk-activated cells39, 40 and the puc enhancer trap line has been widely employed to monitor Bsk activity in vivo.35, 41, 42 As shown in Figure 2Ab, puc-LacZ enhancer trap line is normally expressed in the stalk region of wing imaginal discs (Figure 2Aa).25, 39, 43 The dMLF overexpression driven by en-GAL4 in the posterior region of the wing discs (Figure 2Ba) induced a strong ectopic expression of puc-lacZ in the posterior region of the wing discs (Figure 2Bb). In contrast, ectopic expression of puc-lacZ was not seen in the control anterior region. These results indicate that overexpression of dMLF can promote ectopic activation of the JNK pathway.
Expression of dMLF and puc-LacZ driven by en-GAL4 in wing imaginal discs. (A) Immunostaining of a wing disc from an en-GAL4/+; pucE69/+ fly with anti-dMLF IgG and anti-LacZ IgG. The settings were different from those in (B) in order to visualize the weak signals of endogeneous dMLF in the control strain. (a) Immunostaining with anti-dMLF IgG (magenta). (b) Immunostaining with anti-LacZ IgG (green). (c) Merged image of dMLF and LacZ signals. (d) Phase contrast image of the wing disc. Scale bars, 100 μm. A, anterior; P, posterior. (B) Immunostaining of a wing disc from an en-GAL4, UAS-dMLF/ +;pucE69/+ fly with anti-dMLF IgG and anti-LacZ IgG. (a) Expression of dMLF (magenta) driven by en-GAL4. (b) Ectopic expression of puc (green) detected in the posterior of the wing disc on overexpression of dMLF. (c) Merged image of anti-dMLF and anti-LacZ signals. These results indicate that overexpression of dMLF induces ectopic JNK activation. (d) Phase contrast image of the wing disc. Scale bars, 100 μm.
As noted previously,26 en-GAL4-driven overexpression of dMLF in the wing imaginal discs induced programmed cell death in the posterior region, although restricted in some areas (Figure 3). Altogether, these data indicate that overexpression of dMLF induces apoptosis through JNK activation during wing development, and consequently an atrophied phenotype of the wing posterior is exhibited in adults.
Overexpression of dMLF in the wing imaginal discs induces programmed cell death. (A) Immunostaining of a wing disc from an en-GAL4/+ fly with anti-activated caspase-3 IgG. (a) Activated Caspase-3 is not apparent without overexpression of dMLF. (b) Phase contrast image. Scale bars, 100 μm. A, anterior; P, posterior. (B) Immunostaining of a wing disc from an en-GAL4, UAS-dMLF/+ fly with anti-dMLF IgG, anti-activated caspase-3 IgG. DNA was labeled with DAPI. (a) Expression of dMLF (green) driven by en-GAL4 in wing discs. (b) Activated Caspase-3 (magenta) is present in posterior of wing discs on overexpression of dMLF. (c) Merged image of anti-dMLF and anti-active Caspase-3 signals. (d) Phase contrast image. Scale bars are for 100 μm. (e–g) A higher magnification of the region marked with the square in (Bc). Fragmentation of DNA (blue) by apoptosis via caspase-3. Scale bars, 20 μm.
dMLF binds to genomic regions containing the DRE sequence of the bsk promoter in S2 cells
A direct interaction between dMLF and DREF has been demonstrated by both yeast two-hybrid assay using DREF as a bait and glutathione S-transferase pull-down assays.13 We reported that the DREF-binding consensus, the DRE sequence, 5′-TATCGATA-3′ exists in the bsk promoter region and DREF activates bsk gene transcription.25 Therefore, an attractive hypothesis is that dMLF and DREF could act together to upregulate bsk transcription. We therefore performed ChIP assays of S2 cell extracts immunoprecipitated with anti-dMLF immunoglobulin G (IgG) followed by quantitative real-time PCR using primers that amplify the bsk gene promoter region containing the DRE sequence (Figure 4a, region 1). The 2 kb upstream region from the transcription initiation site of the bsk gene was chosen as a negative control because it does not contain a DRE sequence (Figure 4a, region 2).
(a) Schematic illustration of the DREF-binding consensus sequence in the 5′-flanking region of the bsk gene. Arrowheads show the positions of primers used for the ChIP assays for two genomic regions (region 1, proximal, and region 2, distal). (b) Chromatin immunoprecipitation results. The data shown are derived from quantitative real-time PCR analysis of two genomic regions 1 and 2. Chromatin from S2 cells was immunoprecipitated with either anti-dMLF IgG or control rabbit IgG. The fold different values are for anti-dMLF IgG immunoprecipitated samples compared with the corresponding control rabbit IgG immunoprecipitated sample defined as 1.00. A sample without antibody treatment was also included as a negative control (no antibody column).
Amplification of the region 1 in the immunoprecipitates with an anti-dMLF IgG was 5.81-fold higher than that with the control IgG (Figure 4b). In contrast, no amplification was observed for the far upstream region 2. These results indicate that dMLF binds to the bsk gene promoter region containing the DRE sequence in vivo.
dMLF is required for bsk gene promoter activity in S2 cells
To further examine the requirement of dMLF for bsk gene promoter activity, we carried out dMLF RNA interference (RNAi) experiments in cultured Drosophila S2 cells (Figure 5). Measuring levels of dMLF proteins in S2 cells by western immunoblot analysis confirmed efficient knockdown of the dMLF gene after treatment with dMLFdsRNA (Figure 5b). We conducted transient luciferase expression assays with the wild-type bsk gene promoter-luciferase reporter gene after treating S2 cells with dMLFdsRNA, DREFdsRNA, control YFPdsRNA or no dsRNA (Mock). Treatment of S2 cells with dMLFdsRNA and DREFdsRNA reduced bsk gene promoter activity by 80% and 90%, respectively, whereas control YFPdsRNA treatment exerted no effect (Figure 5c). These results indicate that both DREF and dMLF are required for bsk promoter activity.
(a) Schematic features of the bsk promoter-luciferase fusion plasmid. The DRE-like sequence is indicated. (b) Western immunoblot analysis of S2 cells treated with dMLFdsRNA, YFPdsRNA or no dsRNA (Mock). Proteins were probed with anti-dMLF IgG and anti-α-tubulin IgG. (c) Effects of dMLFdsRNA treatment on bsk gene promoter activity in S2 cells. Mean activities with s.d. from three independent transfections are shown, with the P-value by Welch’s t-test. (d) dMLF and DREF interact in vivo. Extracts of S2 cells were first immunoprecipitated with anti-dMLF IgG followed by immunoblotting with anti-DREF IgG.
dMLF interacts with DREF in vivo
As reported previously, dMLF genetically interacts with DREF and suppresses the rough-eye phenotype induced by overexpression of DREF.13 Therefore, to further examine the relationship between dMLF and DREF in the wing development, we tested whether changing DREF levels could affect the consequences of dMLF overexpression. Thus, transgenic fly lines carrying UAS-DREF, UAS-dref-IR or UAS-GFP (as a control) were crossed with dMLF overexpression lines. Flies with en-GAL4, UAS-dMLF/ UAS-DREF and UAS-dref-IR/+; en-GAL4, UAS-dMLF/+showed lethality (Table 1). However, when the UAS-GFP transgenic line was used for crossing as a control, the flies were viable (Table 1). In addition, although it is reported that UAS-dref-IR strain 15 was lethal when driven by en-GAL4,44 the en-GAL4 derived expression of UAS-DREF or UAS-dref-IR alone under these conditions exerted no effect on viability (Table 1).45 The en-GAL4 driver is expressed very early and throughout development, and thus its use could lead to early development defects before any adult phenotype could be visualized. The synthetic lethality resulting from DREF and dMLF overexpression could simply be because of the fact that overexpression of each protein leads to cell death. The lethal effect of dMLF overexpression when DREF levels are reduced could be due to the trapping by DNA-unbound dMLF of the remaining low amounts of DREF that would thus become unavailable to control the expression of essential genes. In any of the events, these results suggest that dMLF and DREF act together to play a critical role during Drosophila development.
In addition, dMLF and DREF have been shown to interact directly by both yeast two-hybrid assay using DREF and glutathione S-transferase pull-down assays in vitro.13 Therefore, to investigate this interaction between dMLF and DREF in vivo, immunoprecipitation assays were performed using an anti-dMLF IgG with crude lysates of S2 cells. In immunoblot analyses of the immunoprecipitates with anti-dMLF IgG, bands for DREF were detected (Figure 5d, lane 2), whereas none were found in the immunoprecipitates with control IgG or no antibody sample (Figure 5d, lanes 3 and 4). These data support the idea that dMLF and DREF truly interact in vivo to regulate target gene expression.
Discussion
Since the identification of dMLF as a partner of DREF by yeast two-hybrid screen, biological significance of interactions of these two proteins has been a long-term puzzle. In the present study, we demonstrated that dMLF acts with DREF to stimulate bsk promoter activity and consequently activates the JNK pathway. Our present data thus shed light on a dMLF function as a co-activator of the transcription factor DREF. DREF is a major regulator of transcription that has been under intensive studies for more than a decade.22, 46 So far, only two negative regulators of DREF have been identified that inhibit the DNA-binding activity of DREF: dMi-2, a component of chromatin remodeling complex,47 and the transcription factor Distal-less.48 The present work thus also constitutes the first example of a possible co-activator of DREF.
Interestingly, dMLF is known to also interact physically and genetically with Su(fu), a negative regulator of the Gli/Ci transcription factor involving Hh signaling pathway.26 Notably, both DREF and Su(fu) share common interactors. Indeed, Yeast two-hybrid screen using DREF and Su(fu) as bait identified both dMLF and dMi-2.26, 47 It would be interesting to compare chromatin immunoprecipitation (ChIP) sequence data with DREF, Su(fu), dMi-2 and dMLF. In addition, dMLF controls the activity of the transcription factor Lozenge by regulating its stability during development of crystal cells.30 Therefore, dMLF appears to contribute to regulation of transcription factor activity by several distinct mechanisms.
In the present study, we found that dMLF overexpression results in an atrophied wing phenotype associated with apoptosis and activation of the JNK signaling pathway. It has been reported that overexpression of dMLF in the wing imaginal disc induces apoptosis and DNA replication,26 whereas the JNK pathway is known to be involved in apoptosis and cell proliferation.32, 33, 34 Therefore, it is suggested that dMLF induces cell proliferation as well as apoptosis via activation of the JNK pathway. There is recent evidence that the JNK pathway activates the transcriptional co-activator Yorkie (Yki) in the Hippo pathway.49 The latter is well known as a tumor suppressor pathway and it represses expression of the apoptosis inhibitor DIAP1 and the cell cycle regulator Cyclin E via inactivation of the Yki by Warts, resulting in cell cycle arrest and induction of apoptosis.50, 51 Therefore, the JNK pathway activated by dMLF may promote cell proliferation through the Hippo pathway. It should also be noted that DREF activates transcription of both the bsk gene25 and the warts gene.24
In the present study, we found that dMLF binds to the bsk gene promoter region containing the DRE sequence in vivo and is required for activation of the bsk promoter. In addition, we confirmed that the dMLF protein interacts with DREF in vivo using immunoprecipitation assays. Although we could not exclude other possibilities, our data support the notion that dMLF-induced JNK activation is, at least in part, mediated by DREF. Indeed, these results suggest that dMLF forms complexes with DREF and both factors act together to enhance bsk promoter activity and consequently activate the JNK pathway. In addition, we cannot exclude the possibility that dMLF may directly binds to the bsk gene promoter region. However, as dMLF is known to physically interact with DREF,13 it is more likely recruited on the bsk promoter via DREF. Although our findings were obtained with the wing system, JNK pathway regulation by dMLF may also be important in hemocyte differentiation. In any event, dMLF and DREF might control complex signaling networks in a well-balanced way. Identification of additional genetic interactants with dMLF by genetic screening may provide clues for deeper understanding of dMLF functions in vivo.
Materials and methods
Oligonucleotides
To carry out ChIP assays, the following PCR primers were chemically synthesized. These primer sets were designed to amplify 150 base pair (bp) amplicons: bsk+60, F: 5′-GCGGCACTTTCGCATGAGAATAATTG-3′ bsk–90, R: 5′-TCGATTGGCTGACTTTAGCCGTTTCT-3′; bsk–1999, F: 5′-TTCAGGGATATGAACGCAAATTGCCG-3′; bsk–2149, R: 5′-AATGCTGACGTTCTTCAGTTGCTTGG-3′.
Plasmids
The plasmids p5′-1000bskwt-Luc and p5′-1000bskmutDRE-Luc used in the luciferase transient expression assays were as described previously.25, 43
Fly stocks
Flies were cultured at 18 °C or 25 °C on standard food. Canton S flies were used as the wild-type strain. The UAS-dMLF(II) line has been described previously26 and the en-GAL4 driver line was kindly provided by Dr N Dyson (Harvard Medical School, Charlestown, MA, USA). The pucE69/TM3 line was kindly provided by Dr T Adachi-Yamada (Gakushuin University, Tokyo, Japan). The mutant stocks bsk1/CyO and bsk2/CyO used in this study were obtained from the Bloomington Drosophila stock center (Bloomington, IN, USA). The UAS-DREF (II) and UAS-dref-IR (X) lines were as described previously.44
Inspection of wing phenotype
Ripped wings of adult flies were mounted on slides with Hoyer’s medium and inspected with an Olympus SZX12 microscope equipped with an Olympus CAMEDIA C-3030 ZOOM (Olympus, Tokyo, Japan).
Immunohistochemistry
Third instar larvae were dissected in Drosophila Ringer’s solution and imaginal discs were collected and fixed in 4% paraformaldehyde in phosphate-buffered saline (PBS) for 15 min at 25 °C. After washing with PBS containing 0.3% Triton X-100 (PBS-T), the samples were blocked with PBS-T containing 10% normal goat serum for 30 min at 25 °C and incubated with rabbit anti-dMLF antibody (1:500), rat anti-dMLF polyclonal antibody (1:500), mouse anti-β-galactosidase monoclonal antibody (1:500) or rabbit anti-active caspase-3 antibody (1:500; BD Pharmingen, Tokyo, Japan) at 4 °C for 16 h. After extensive washing with PBS-T, the imaginal discs were incubated with anti-rabbit IgG conjugated with Alexa 488 (1:400; Invitrogen, Life Technologies, San Diego, CA, USA) or anti-mouse IgG conjugated with Alexa 546 (1:400; Invitrogen) for 3 h at 25 °C. After extensive washing with PBS-T and PBS, samples were mounted in Vectashield (Vector Laboratories Inc., Burlingame, CA, USA) and inspected with an Olympus FLUOVIEW FV10i.
Preparation of double-stranded RNA (dsRNA) for RNAi experiments
The dsRNA was prepared using a RiboMax T7 kit (Promega, Madison, WI, USA) according to the manufacturer’s instructions. RNAi analysis was carried out as described earlier.21, 52
Chromatin immunoprecipitation
ChIP was performed using a ChIP Assay kit as recommended by the manufacturer (Merck Millipore, Billerica, MA, USA) with minor modifications.53 Approximately 2 × 107 S2 cells were fixed in 1% formaldehyde at 37 °C for 10 min, quenched in 125 mM glycine for 5 min at 25 °C, collected and washed twice in PBS containing protease inhibitors (1 mM PMSF, 1 μg/ml aprotinin and 1 μg/ml pepstatin A) and then lysed in 2 ml of SDS lysis buffer (Merck Millipore). Lysates were sonicated to break DNA into fragments of <1 kb and centrifuged at 15 300 g for 10 min at 4 °C. The sonicated cell supernatants were diluted 10-fold in ChIP Dilution Buffer (Merck Millipore) and precleared with 80 μl of salmon sperm DNA/protein A agarose-50% slurry for 30 min at 4 °C. After brief centrifugation, supernatants were incubated with 1 μg of normal rabbit IgG (Sigma-Aldrich, St Louis, MO, USA) or anti-dMLF rabbit IgG for 16 h at 4 °C. Salmon sperm DNA/protein A agarose-50% slurry was added, followed by incubation for 1 h at 4 °C. After washing, immunoprecipitated DNA was eluted with elution buffer containing 1% SDS and 0.1 M NaHCO3. Then, protein–DNA crosslinks were reversed by heating at 65 °C for 4 h. After deproteinization with proteinase K, DNA was recovered. Then, the immunoprecipitated DNA fragments were detected by quantitative real-time PCR using SYBR Green I (Takara Bio Inc., Shiga, Japan) and the Applied Biosystems 7500 Real Time PCR system (Life Technologies, Foster City, CA, USA). The ΔΔCT value of each sample was calculated by subtracting the CT value for the input sample from the CT value obtained for the immunoprecipitated sample. Fold change was calculated by raising 2 to the ΔΔCT power. The ΔΔCT was calculated by subtracting the ΔCT value for the sample immunoprecipitated with control IgG.54
Luciferase transient expression assays
For luciferase transient expression assays, 1 × 105 S2 cells were plated in 24-well dishes. Transfection of various DNA mixtures was performed using Cell-Fectin reagent (Invitrogen) and cells were harvested 48 h thereafter. Luciferase activity was measured as described earlier21, 52, 55 and normalized to Renilla luciferase activity using pAct5C-seapansy as an internal control.18 All plasmids for transfection were prepared using a QIAGEN plasmid Kit (Qiagen, Venlo, Netherlands).
For dsRNA interference experiments, 30 μg of dMLFdsRNA, DREFdsRNA or YFPdsRNA were added to 1 × 106 S2 cells plated in each of six-well dishes. At 72 h after RNAi treatment, the cells were transfected with various DNA mixtures and harvested 48 h later for processing for the luciferase assay as described above.
All transient expression data reported in this study are means from three independent experiments, each performed in triplicate. Average relative luciferase activity was graphed and statistically analyzed with the Welch’s t-test.
Western immunoblot analysis
Whole-cell extracts from S2 cells, prepared as described earlier,43 were applied to 10% polyacrylamide gels containing 0.1% SDS and transferred to polyvinylidene difluoride membranes. Blotted membranes were blocked with Tris-buffered saline (50 mM Tris–HCl, pH 8.3 and 150 mM NaCl) containing 10% skim milk for 1 h at 25 °C and incubated with an anti-dMLF IgG in a 1:1000 dilution, or an anti-α tubulin monoclonal antibody (Sigma-Aldrich) in a 1:5000 dilution at 4 °C for 16 h. After washing with Tris-buffered saline, the blots were incubated with horseradish peroxidase-conjugated anti-rabbit IgG or horseradish peroxidase-conjugated anti-mouse IgG (Life Technologies, Waltham, MA, USA) in a 1:5000 dilution for 1 h at 25 °C. Detection was performed with ECL Western blotting detection reagents (GE Healthcare, Buckinghamshire, UK), and images were analyzed with a Lumivision Pro HSII image analyzer (Aisin Seiki, Aichi, Japan).
Immunoprecipitation
Whole-cell extracts were prepared in lysis buffer containing a proteinase inhibitor mix. Immunoprecipitation was performed with 800 μg of lysates with anti-dMLF rabbit IgG or the normal rabbit IgG (Sigma-Aldrich) and protein A–Sepharose (GE Healthcare). The protein A–antibody complexes were washed with the same buffer. Immunoprecipitates were separated by SDS–PAGE and immunoblotted with mouse anti-DREF IgG.15
References
Yoneda-Kato N, Look AT, Kirstein MN, Valentine MB, Raimondi SC, Cohen KJ et al. The t(3;5)(q25.1;q34) of myelodysplastic syndrome and acute myeloid leukemia produces a novel fusion gene, NPM-MLF1. Oncogene 1996; 12: 265–275.
Feuerstein N, Spiegel S, Mond JJ . The nuclear matrix protein, numatrin (B23), is associated with growth factor-induced mitogenesis in Swiss 3T3 fibroblasts and with T lymphocyte proliferation stimulated by lectins and anti-T cell antigen receptor antibody. J Cell Biol 1988; 107: 1629–1642.
Chan PK . Cross-linkage of nucleophosmin in tumor cells by nitrogen mustard. Cancer Res 1989; 49: 3271–3275.
Border RA, Lehner CF, Eppenberger HM, Nigg EA . Major nucleolar proteins shuttle between nucleus and cytoplasm. Cell 1989; 56: 379–390.
Muslin AJ, Tanner JW, Allen PM, Shaw AS . Interaction of 14-3-3 with signaling proteins is mediated by the recognition of phosphoserine. Cell 1996; 84: 889–897.
Yaffe MB, Rittinger K, Volinia S, Caron PR, Aitken A, Leffers H et al. The structural basis for 14-3-3: phosphopeptide binding specificity. Cell 1997; 91: 961–971.
Lim R, Winteringham LN, Williams JH, McCulloch RK, Ingley E, JY-H Tiao et al. MADM a novel adaptor protein that mediates phosphorylation of the 14-3-3 binding site of myeloid leukemia factor 1. J Biol Chem 2002; 277: 40997–41008.
Hanissian SH, Akbar U, Teng B, Janjetovic Z, Hoffmann A, Hitzler JK et al. cDNA cloning and characterization of a novel gene encoding the MLF1-interacting protein MLF1IP. Oncogene 2004; 23: 3700–3707.
Yoneda-Kato N, Tomoda K, Umehara M, Arata Y, Kato J-Y. . 2005. Myeloid leukemia factor 1 regulates p53 by suppressing COP1 via COP9 signalosome subunit 3. EMBO J 2005; 24: 1739–1749.
Winteringham LN, Endersby R, Kobelke S, McCulloch RK, Williams JH, Stillitano J et al. Myeloid leukemia factor 1 associates with a novel heterogeneous nuclear ribonucleoprotein U-like molecule. J Biol Chem 2006; 281: 38791–38800.
Kuefer MU, Look AT, Williams DC, Valentine V, Naeve CW, Behm FG et al. cDNA cloning, tissue distribution, and chromosomal localization of myelodysplasia/myeloid leukemia factor 2 (MLF2). Genomics 1996; 35: 392–396.
Williams JH, Daly LN, Ingley E, Beaumont JG, Tilbrook PA, Lalonde J-P et al. HLS7, a hemopoietic lineage switch gene homologous to the leukemia-inducing gene MLF1. EMBO J 1999; 18: 5559–5566.
Ohno K, Takahashi Y, Hirose F, Inoue Y-H, Taguchi O, Nishida Y et al. Characterization of a Drosophila homologue of the human myelodysplasia/myeloid leukemia factor (MLF). Gene 2000; 260: 133–143.
Hirose F, Yamaguchi M, Hamada H, Inomata Y, Matsukage A . Novel 8-base pair sequence (Drosophila DNA replication-related element) and specific binding factor involved in the expression of Drosophila genes for DNA polymerase alpha and proliferating cell nuclear antigen. J Biol Chem 1993; 268: 2092–2099.
Hirose F, Yamaguchi M, Kuroda K, Omori A, Hachiya T, Ikeda M et al. Isolation and characterization of cDNA for DREF, a promoter-activating factor for Drosophila DNA replication- related genes. J Biol Chem 1996; 271: 3930–3937.
Ohno K, Hirose F, Sakaguchi K, Nishida Y, Matsukage A . Transcriptional regulation of the Drosophila Cyc A gene by the DNA replication-related element (DRE) and DRE binding factor (DREF). Nucleic Acids Res 1996; 24: 3942–3946.
Takahashi Y, Yamaguchi M, Hirose F, Cotterill S, Kobayashi J, Miyajima S et al. DNA replication-related elements cooperate to enhance promoter activity of the Drosophila DNA polymerase α 73-kDa subunit gene. J Biol Chem 1996; 271: 14541–14547.
Sawado T, Hirose F, Takahashi Y, Sasaki T, Shinomiya T, Sakaguchi K et al. The DNA replication-related element (DRE) / DRE-binding factor system is a transcriptional regulator of the Drosophila E2F gene. J Biol Chem 1998; 273: 26042–26051.
Okudaira K, Ohno K, Yoshida H, Asano M, Hirose F . Transcriptional regulation of the Drosophipla orc2 gene by the DREF pathway. Biochim Biophys Acta 2005; 1732: 23–30.
Tsuchiya A, Inoue YH, Ida H, Kawase Y, Okudaira K, Ohno K et al. Transcriptional reguration of the Drosophila rfc1 gene by the DRE-DREF pathway. FEBS J 2007; 274: 1818–1832.
Ida H, Yoshida H, Nakamura K, Yamaguchi M . Identification of the Drosophila eIF4A gene as a target of the DREF transcription factor. Exp Cell Res 2007; 313: 4208–4220.
Matsukage A, Hirose F, Yoo MA, Yamaguchi M . The DRE/DREF transcriptional regulatory system: a master key for cell proliferation. Biochim Biophys Acta 2008; 1779: 81–89.
Nakamura K, Ida H, Yamaguchi M . Transcriptional regulation of the Drosophila moira and osa genes by the DREF pathway. Nucleic Acids Res 2008; 36: 3905–3915.
Fujiwara S, Ida H, Yoshioka Y, Yoshida H, Yamaguchi M . The warts gene as a novel target of the Drosophila DRE / DREF transcription pathway. Am J Cancer Res 2012; 2: 35–44.
Yoshioka Y, Trong-Tue N, Fujiwara S, Matsuda R, Valadez-Graham V, Zurita M et al. Drosophila DREF acting via the JNK pathway is required for thorax development. Genesis 2012; 50: 599–611.
Fouix S, Martin-Lannerée S, Sanial M, Morla L, Lamour-Isnard C, Plessis A . Over-expression of a novel nuclear interactor of suppressor of fused, the Drosophila myelodysplasia/myeloid leukemia factor, induces abnormal morphogenesis associated withincreased apoptosis and DNA synthesis. Genes Cells 2003; 8: 897–911.
Sugano W, Ohno K, Yoneda-Kato N, Kato JY, Yamaguchi M . The myeloid leukemia factor interacts with COP9 signalosome subunit 3 in Drosophila melanogaster. FEBS J 2008; 275: 588–600.
Kazemi-Esfarjani P, Benzer S . Suppression of polyglutamine toxicity by a Drosophila homolog of myeloid leukemia factor 1. Hum Mol Genet 2002; 11: 2657–2672.
Kim WY, Fayazi Z, Bao X, Higgins D, Kazemi-Esfarjani P . Evidence for sequestration of polyglutamine inclusions by Drosophila myeloid leukemia factor. Mol Cell Neurosci 2005; 29: 536–544.
Bras S, Martin-Lanneree S, Gobert V, Auge B, Breig O, Sanial M et al. MLF is a conserved regulator of RUNX transcription factor activity involved in hematopoiesis. Proc Natl Acad Sci USA 2012; 109: 4986–4991.
Martin-Blanco E, Pastor-Pareja JC, Garcia-Bellido A . JNK and decapentaplegic signaling control adhesiveness and cytoskeleton dynamics during thorax closure in Drosophila. Proc Natl Acad Sci USA 2000; 97: 7888–7893.
Noselli S, Aqnes F . Roles of the JNK signaling pathway in Drosophila morphogenesis. Curr Opin Genet Dev 1999; 9: 466–472.
Davis RJ . Signal transduction by the JNK group of MAP kinases. Cell 2000; 13: 239–252.
Igaki T, Paqliarini RA, Xu T . Loss of cell polarity drives tumor growth and invasion through JNK activation in Drosophila. Curr Biol 2006; 6: 1139–1146.
Adachi-Yamada T, Fujimura-Kamada K, Nishida Y, Matsumoto K . Distortion of proximodistal information causes JNK-dependent apoptosis in Drosophila wing. Nature 1999; 400: 166–169.
Riesgo-Escovar JR, Jenni M, Fritz A, Hafen E . The Drosophila Jun-N-terminal kinase is required for cell morphogenesis but not for DJun-dependent cell fate specification in the eye. Genes Dev 1996; 10: 2759–2768.
Sluss HK, Han Z, Barrett T, Davis RJ, Ip YT . AJNK signal transduction pathway that mediates morphogenesis and an immune response in Drosophila. Genes Dev 1996; 10: 2745–2758.
Agnes F, Suzanne M, Noselli S . The Drosophila JNK pathway controls the morphogenesis of imaginal discs during metamorphosis. Development 1999; 126: 5453–5462.
Adachi-Yamada T, O’Connor MB . Morphogenetic apoptosis: a mechanism for correcting discontinuities in morphogen gradients. Dev Biol 2002; 251: 74–90.
Martin-Blanco E, Gampel A, Ring J, Virdee K, Kirov N, Tolkovsky AM et al. Puckered encodes a phosphatase that mediates a feedback loop regulating JNK activity during dorsal closure in Drosophila. Genes Dev 1998; 12: 557–570.
Tateno M, Nishida Y, Adachi-Yamada T . Regulation of JNK by Src during Drosophila development. Science 2000; 287: 324–327.
Igaki T, Kanda H, Yamamoto-Goto Y, Kanuka H, Kuranaga E, Aigaki T et al. Eiger a TNF superfamily ligand that triggers the Drosophila JNK pathway. EMBO J 2002; 21: 3009–3018.
Yoshioka Y, Suyari O, Yamaguchi M . Transcription factor NF-Y is involved in regulation of the JNK pathway during Drosophila thorax development. Genes Cells 2008; 13: 117–130.
Yoshida H, Kwon E, Hirose F, Otsuki K, Yamada M, Yamaguchi M . DREF is required for EGFR signaling during Drosophila wing vein development. Genes Cells 2004; 9: 935–944.
Yoshida H, Inoue YH, Hirose F, Sakaguchi K, Matsukage A, Yamaguchi M . Over-expression of DREF in the Drosophila wing imaginal disc induces apoptosis and a notching wing phenotype. Genes Cells 2001; 6: 877–886.
Fujiwara S, Yoshida H, Yamaguchi M . DREF a concertmaster for Hippo pathway and JNK pathway in Drosophila. J Carcinogene Mutagene 2013; 4: e110.
Hirose F, Ohshima N, Kwon EJ, Yoshida H, Yamaguchi M . Drosophila Mi-2 negatively regulates dDREF by inhibiting its DNA-binding activity. Mol Cell Biol 2002; 22: 5182–5193.
Hayashi Y, Kato M, Seto H, Yamaguchi M . Drosophila Distal-less negatively regulates dDREF by inhibiting its DNA binding activity. Biochim Biophys Acta 2006; 1759: 359–366.
Sun G, Irvine KD . Regulation of Hippo signaling by Jun kinase signaling during compensatory cell proliferation and regeneration, and in neoplastic tumors. Dev Biol 2011; 350: 139–151.
Xu T, Wang W, Zhang S, Stewart RA, Yu W . The Drosophila tumor suppressor gene warts encodes a homolog of human myotonic dystrophy kinase and is required for the control of cell shape and proliferation. Genes Dev 1995; 9: 534–546.
Huang J, Wu S, Barrera J, Matthews K, Pan D . The Hippo signaling pathway coordinately regulates cell proliferation and apoptosis by inactivating Yorkie, the Drosophila homolog of YAP. Cell 2005; 122: 421–434.
Seto H, Hayashi Y, Kwon E, Taguchi O, Yamaguchi M . Antagonistic regulation of the Drosophila PCNA gene promoter by DREF and Cut. Genes Cells 2006; 11: 499–512.
Thao DTP, Ida H, Yoshida H, Yamaguchi M . Identification of Drosophila skpA gene as novel target of the transcription factor DREF. Exp Cell Res 2006; 312: 3641–3650.
Morrison T, Weis JJ, Wittwer CT . Quantification of low-copy transcripts by continuous SYBR Green I monitoring during amplification. Biotechniques 1998; 24: 954–962.
Hayashi Y, Yamagishi M, Nishimoto Y, Taguchi O, Matsukage A, Yamaguchi M . A binding site for the transcription factor Grainyhead/Nuclear transcription factor-1 contributes to regulation of the Drosophila proliferating cell nuclear antigen gene promoter. J Biol Chem 1999; 274: 35080–35088.
Acknowledgements
This study was partially supported by Grants-in-Aid from JSPS, JST and the Ministry of Education, Science, Sports and Culture of Japan as well as by the French ARC. We thank Dr T Adachi-Yamada and Dr N Dyson for fly strains and Dr M Moore for comments in English usage.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 3.0 Unported License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/3.0/
About this article
Cite this article
Yanai, H., Yoshioka, Y., Yoshida, H. et al. Drosophila myeloid leukemia factor acts with DREF to activate the JNK signaling pathway. Oncogenesis 3, e98 (2014). https://doi.org/10.1038/oncsis.2014.13
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/oncsis.2014.13
Keywords
This article is cited by
-
The Hippo pathway as a target of the Drosophila DRE/DREF transcriptional regulatory pathway
Scientific Reports (2014)