Abstract
Kaposi's sarcoma herpesvirus (KSHV) latent oncoprotein viral FLICE (FADD-like interferon converting enzyme)-like inhibitory protein (v-FLIP) or K13, a potent activator of NF-κB, has well-established roles in KSHV latency and oncogenesis. KSHV-induced COX-2 represents a novel strategy employed by KSHV to promote latency and inflammation/angiogenesis/invasion. Here, we demonstrate that v-FLIP/K13 promotes tumorigenic effects via the induction of host protein COX-2 and its inflammatory metabolite PGE2 in an NF-κB-dependent manner. In addition to our previous studies demonstrating COX-2/PGE2's role in transcriptional regulation of KSHV latency promoter and latent gene expression, the current study adds to the complexity that though LANA-1 (latency associated nuclear antigen) is utilizing COX-2/PGE2 as critical factors for its transcriptional regulation, it is the v-FLIP/K13 gene in the KSHV latency cluster that maintains continuous COX-2/PGE2 levels in the infected cells. We demonstrate that COX-2 inhibition, via its chemical inhibitors (NS-398 or celecoxib), reduced v-FLIP/K13-mediated NF-κB induction, and extracellular matrix (ECM) interaction-mediated signaling, mitochondrial antioxidant enzyme manganese superoxide dismutase (MnSOD) levels, and subsequently downregulated detachment-induced apoptosis (anoikis) resistance. vFLIP expression mediated the secretion of cytokines, and spindle cell differentiation activated the phosphorylation of p38, RSK, FAK, Src, Akt and Rac1-GTPase. The COX-2 inhibition in v-FLIP/K13-HMVECs reduced inflammation and invasion/metastasis-related genes, along with reduced anchorage-independent colony formation via modulating ‘extrinsic’ as well as ‘intrinsic’ cell death pathways. COX-2 blockade in v-FLIP/K13-HMVEC cells drastically augmented cell death induced by removal of essential growth/survival factors secreted in the microenvironment. Transformed cells obtained from anchorage-independent colonies of COX-2 inhibitor-treated v-FLIP/K13-HMVEC cells expressed lower levels of endothelial–mesenchymal transition genes such as slug, snail and twist, and higher expression of the tumor-suppressor gene, E-cadherin. Taken together, our study provides strong evidences that FDA-approved COX-2 inhibitors have great potential in blocking tumorigenic events linked to KSHV's oncogenic protein v-FLIP/K13.
Similar content being viewed by others
Introduction
Human herpesvirus-8/Kaposi's sarcoma herpesvirus (KSHV) is associated with a neoplastic angioproliferative endothelial malignancy called Kaposi sarcoma (KS) and two other AIDS-related lymphoid cell malignancies called primary effusion lymphoma and multicentric Castleman's disease.1, 2 Endothelial tumors associated with KSHV infection are unique as these not only resemble chronic inflammation but also present cells unlike classical tumor cells with a spindle phenotype. Tumor cells in KS lesions are ‘cytokine-driven’ and are essentially dependent upon paracrine growth signals in the form of cytokines such as IL-6, IL-1β, tumor necrosis factor-α (TNF-α), interferon-γ, basic fibroblast growth factor and vascular endothelial growth factor (VEGF) secreted from the infected cells. In late stage nodular KS lesions, >90% of the spindle cells contain latent KSHV3, 4 and express viral FLICE (FADD-like interferon converting enzyme)-like inhibitory protein (v-FLIP) as one of a limited number of latent proteins.1, 2 The viral life cycle displays distinct latent and lytic replication events, where infected cell survival and proliferation is exclusively dependent on KSHV latency. Previous studies from our lab have shown that KSHV utilizes COX-2/PGE2 to maintain its latency and pathogenesis, and COX-2/PGE2 are regulated at the transcriptional level in de novo KSHV-infected cells.5, 6, 7, 8, 9 We hypothesized that the sustained action of COX-2/PGE2 might be a function of one of the viral latent proteins, and targeting indicated that KSHV protein could be an effective therapeutic strategy against KSHV-associated malignancies.
v-FLIP has been shown to perform multiple functions, including upregulation of inflammatory cytokines IL-8 and IL-6, induction of master signal cascade regulator molecule NF-κB, spindling phenotype in infected ECs and regulation of inflammation, and cell proliferation and immune responses.10, 11 v-FLIP has been shown to induce COX2 in previous studies12, 13 but it has never been studied in detail for the downstream functions of COX-2/PGE2 and the potential/efficacy of COX-2 inhibitors in controlling v-FLIP-induced oncogenesis. KS progression has been linked to a number of critical events such as overcoming the requirement for the extracellular matrix (ECM; a complex meshwork of macromolecules, such as fibronectin, vitronectin, laminin and collagen) for growth, evading anoikis, altering the biological repertoire of the ECs and metastasizing to different distant organs. Anoikis, meaning loss of ‘home’ or ‘homelessness,’ originally defined as a unique phenomenon reflecting apoptotic cell death consequential to inadequate/insufficient/inappropriate ECM interactions14 or suspension-induced apoptosis, is an essential mechanism for maintaining the correct position of cells within tissues and is recognized as a potentially significant factor in tumor angiogenesis and metastasis.14 v-FLIP has been shown to inhibit anoikis of primary endothelial cells15 and COX-2/PGE2 have been reported to have important roles in regulating anoikis in many cancers.16 Therefore, we planned to explore the mechanisms by which v-FLIP-induced COX-2/PGE2 participate in breaching anoikis, deregulating infected cell–ECM interactions and impairing apoptosis of infected cells, thereby contributing to oncogenesis. To understand the role of COX-2/PGE2, we utilized two inhibitors of COX-2, NS-398 and celecoxib. NS-398 (N-(2-cyclohexyloxy-4-nitrophenyl)-methanesulfonamide) is a COX-2-specific inhibitor that has been shown to have chemotherapeutic potential against colon and pancreatic cancer cells. Celecoxib has demonstrated its chemotherapeutic properties in a variety of cancers including colon, breast, skin, prostate and pancreatic cancer cells, but has never been tested in KSHV-associated malignancies.
Collectively, these studies show the interplay between vFLIP and COX-2. We demonstrate that vFLIP activates COX-2/PGE2 in a NF-κB-dependent manner and conversely COX-2/PGE2 is required for vFLIP-induced NF-κB activation, ECM interaction, FAK/Src/AKT, Rac1 activation, mitochondrial antioxidant enzyme manganese superoxide dismutase (MnSOD) level and anokises resistance. Taken together, our results present the less explored clinical perspective of COX-2 inhibitors (celecoxib and NS-398) in controlling inflammation-related cytokines, anoikis resistance, ECM interaction-induced signaling events, cell adhesion, anchorage-resistant colony formation, modulation of MnSOD and endothelial–mesenchymal transitions (EndMTs) induced by v-FLIP. Impaired apoptosis is a hallmark of malignancies that underpins both oncogenesis and resistance to chemotherapies, and the ultimate aim of cancer treatment is to inhibit the growth of precancerous and cancerous cells without affecting the normal cells. Along with the very encouraging data from our ongoing in vivo studies with a panel of COX-2 inhibitors, the current study holds significant impact on the design of therapies against KSHV-associated neoplasia and sheds light on the underlining events of KS pathogenesis.
Results
Relationship between v-FLIP, NF-κB, MAPKs and COX-2
KSHV latency gene v-FLIP induces COX-2, mPGES-1 gene expression, COX-2 protein levels and PGE2 secretion
As our extensive study showed a tight connection between inflammation-associated stress response gene COX-2 and its inflammatory metabolite PGE2 during KSHV latent infection, we wanted to investigate which viral latent protein (s) preferentially induces this inflammatory pathway. KSHV latency genes (LANA-1, v-FLIP, v-cyclin, K8 and K12) were transduced (∼90% efficiency; Supplementary Figure S1Aa and b) in HMVEC-d cells using a lentiviral transduction method. Successful transduction of viral genes was verified by quantitating the expression of specific viral genes (data not shown). A higher percentage of v-FLIP/K13-HMVECs exhibited the characteristic elongated spindle phenotype (Supplementary Figure S1Ad) as compared to the cobblestone morphology of untransduced or p-SIN-HMVEC cells (Supplementary Figure S1Ac). v-FLIP/K13-HMVEC cells stained positive for anti-v-FLIP/K13 (Supplementary Figure S1B) and exhibited increased COX-2 (Figure 1a) and mPGES-1 (Figure 1b) gene expression and PGE2 secretion (Figure 1c) without any effect on COX-1 (data not shown). While examining COX-2 protein levels from the lysates prepared from these cells, only v-FLIP-transduced cells showed elevated COX-2 protein levels (Figure 1d) without affecting COX-1 (Figure 1d). Cells infected with the lentivirus-expressing GFP did not induce COX-2, confirming the specificity of induction (Figures 1a–d).
Effect of v-FLIP/K13 on COX-2, mPGES-1 gene expression, COX-2 and COX-1 protein levels and PGE2 secretion and role of cis-acting factors on transcriptional regulation of COX-2 promoter by v-FLIP/K13. (a, b) HMVEC-d cells were infected with KSHV latency gene (LANA-1, v-FLIP, v-Cyclin, K8 and K12), GFP or empty vector (pSIN) expressing lentiviruses. Forty-eight hours after infection, cells were washed, lysed, total RNA was prepared, and COX-2 and mPGES-1 expression were quantitated using COX-2 and mPGES-1 gene-specific primers normalized to GAPDH levels as described earlier.5 Fold induction was calculated by considering levels in pSIN lentivirus-infected cells as onefold. (c) Supernatants collected from cells in (a) were used for PGE2 detection by methods described previously.5 Each reaction was done in triplicate and each point represents the average±s.d. of three independent experiments. (d) Lysates prepared from the above mentioned cells were used for quantitating COX-2 protein levels. (e) A total of 293 cells seeded in 24-well tissue culture plate were fed with antibiotic-free low serum (0.5% FBS) Dulbecco's modified Eagle's medium for 18 h before transfection using Lipofectamine 2000 (Invitrogen). Low serum conditions were maintained throughout the experiment. In all, 293 cells were transfected with COX-2 full-length promoter construct (P2-1900-luc), along with increasing concentrations (0.5–3 μg) of either control pcDNA or v-FLIP/K13 expression plasmid. Forty-eight hours after transfection, cells were harvested, lysed and assayed for R.L.U. as described before. Promoter induction in control pcDNA cotransfected cells was considered as onefold. (f) A total of 293 cells were cotransfected with various COX-2 deletion constructs (as mentioned in the supplement) along with 2 μg each of control pcDNA or v-FLIP/K13 expression plasmid. Forty-eight hours after transfection, cells were harvested and assayed for R.L.U. as described earlier. (g) 293 cells were cotransfected with COX-2 promoter constructs (P2-1900 or P2-1102) along with 2 μg expression plasmids of pcDNA, IKK2DN, IκβM, p65 or VIVIT. Promoter induction in v-FLIP/K13-transfected cells was considered as 100%. Data in Figure 1E–G represent the mean R.L.U. after normalizing with the cotransfected Renilla activity. Each reaction was done in triplicate and each point represents the average±s.d. of three experiments. Promoter induction in pcDNA-transfected cells was considered as onefold. (h) pSIN or v-FLIP/K13-transduced HMVEC-d cells were untreated or treated with either DMSO solvent control or NF-κB inhibitor Bay 11-7082 for 1 h. Inhibitor or solvent control were removed after 1 h and cells were replenished with growth medium for 24 h or 48 h and COX-2 gene expression was measured. COX-2 gene expression in pSIN-HMVECs was considered as onefold. COX-2 gene expression in untreated v-FLIP/K13-HMVECs was considered as 100%. (i) v-FLIP transduction in HMVECs induces NFAT activation. Relative activation was calculated by taking the activation in pSIN-transduced cells as onefold at all of the indicated times. (j) v-FLIP transduction in HMVECs induces CREB activation (CREB serine 133 phosphorylation). Relative activation was calculated by taking the activation in p-SIN-HMVECs as onefold at the indicated times. * denotes statistically significant at P<0.01. Red arrows pointing down denote decrease.
v-FLIP/K13 modulates COX-2 transcription via NF-κB and CRE cis-acting elements in the human COX-2 promoter
In order to understand whether v-FLIP/K13 induced COX-2/PGE2 at the transcriptional level, we performed COX-2 promoter analysis in the presence of v-FLIP/K13 expression plasmid. At 2 μg of v-FLIP, maximum (sixfold) induction in COX-2 gene expression was observed (Figure 1e) with no effect on the COX-1 promoter (P2-1009) reaffirming the specificity for COX-2 induction (data not shown). Along with our previous study showing the important role of NFAT and CREB in KSHV-induced COX-2, many other transcription factors such as NF-κB, nuclear factor-IL-6, CRE and AP-1 have also been shown to regulate COX-2 transcription in other systems.17 We sought to assess the transcription factors involved in v-FLIP-induced COX-2 using various deletion constructs of COX-2 (Supplementary Figure S1C). v-FLIP transduction induced the activity of the COX-2 promoter luciferase construct (Figure 1f). Loss of one NF-κB site (P2-431) resulted in 77% inhibition in COX-2 promoter activity, whereas loss of both NF-κB (P2-274) sites increased the inhibition to 88%, confirming the exclusive role of NF-κB (Figure 1f). An 88% reduction was also observed in the construct without NF-κB, AP2/SP1, NF-IL6 and dNFAT (P2-192) (Figure 1f). Removal of pNFAT and AP-1 enhanced the COX-2 promoter activity, suggesting a role of CRE in modulating the v-FLIP-mediated COX-2 transcription (Figure 1f). To verify the role of NF-κB in transcriptional regulation of v-FLIP-induced COX-2, we measured COX-2 promoter activity in the presence of 1 μg each of p65, IKK2DN, IκBM (dominant negative mutant IκB blocking NF-κB activation) and VIVIT expression plasmids (Figure 1g). Cotransfection with either IκBM or IKK2DN effectively inhibited COX-2 promoter activation (Figure 1g). The role of NF-κB was also confirmed by assessing the effect of NF-κB inhibitor Bay 11-7082 on v-FLIP-HMVEC cells. We observed a 57% and 68% reduction in COX-2 gene expression at 24 h and 48 h, respectively (Figure 1h). As complete inhibition of v-FLIP-mediated COX-2 promoter activity and gene expression was not obtained by NF-κB inhibition, we reasoned that other transcription factors also regulate v-FLIP-induced COX-2. COX-2 luciferase reporter activity was reduced by 40% in the presence of a plasmid expressing the peptide VIVIT, suggesting some role of NFAT activation in v-FLIP-mediated COX-2 activity (Figure 1g). As KSHV infection-induced COX-2 transcription is regulated via NFAT and CREB, we checked whether these factors are involved in v-FLIP-mediated COX-2 activity or even if v-FLIP transduction induced the activity of NFAT and CREB. We observed more than twofold induction in NFAT and CREB as indicated in Figures 1i and j, respectively.
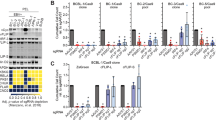
v-FLIP/K13-induced multiple MAPKs regulate COX-2 gene expression, protein levels and PGE2 secretion
COX-2 promoter analysis indicated a role for CRE (Figure 1f), and v-FLIP-transduced cells showed higher activation of CREB at 24 h and 48 h of transduction (Figure 1j). CREB is a member of a large family of structurally related transcription factors (AFT 1 to -4, c-Fos, c-Jun, c-Myc and C/EBP), which specifically recognizes the CRE promoter site (5′-TGACGTCA-3′). CREB activity is exclusively controlled by a variety of MAPKs that phosphorylate CREB at Ser-133, which increases CREB's affinity for its promoter site. As many MAPKs have been shown to regulate CREB activity, we followed to assess MAPK activity upon v-FLIP/K13 transduction. MAPK activation was analyzed using phospho-MAPK arrays (Figure 2a, Supplementary Figures S2A and S2B) as per the methods described previously.8 Among all the MAPKs tested (Figure 2b), v-FLIP/K13 induced the phosphorylation of P38 (-γ), P38 (–δ), RSK (1), RSK (2), GSK-3 (α/β), AKT (1), and AKTpan by 2-, 2.5-, 5.3-, 1-, 3.9-, 6.9- and 6-fold, respectively (Figure 2b). Phosphorylated kinases were also detected in the lysates prepared from p-SIN and v-FLIP/K13-HMVECs (Figure 2c). We observed ERK (2.8), AKT (4.3), P-65 (3.8), RSK1 (4.3) and GSK-3ß (2.4) fold phosphorylation (Figure 2c). To define the role of v-FLIP/K13-induced MAPKs in COX-2 regulation, v-FLIP/K13-HMVECs were untreated or treated with signal cascade (ERK, AKT, P-65, RSK1, and GSK-3ß) inhibitors, in order to quantitate COX-2 gene expression (Figure 2d) and protein levels (Figure 2e). Activation of AKT and GSK3 phosphorylation was also assayed by Fast-activated cell-based ELISA (FACE In-cell Western) (data not shown). Supernatants collected from these cells were assayed for PGE2 (Figure 2f). MEK-inhibitor, U0126, and GSK3–inhibitor, SB216763, did not noticeably inhibit COX-2 gene expression, protein levels or PGE2 secretion as indicated (Figures 2d and f). Pretreatment with P38 inhibitor (SB203580), RSK inhibitor (SL 0101-1) or AKT-inhibitor (API-2) reduced the expression of the COX-2 gene and also the secretion of PGE2, as indicated (Figures 2d and f). Pretreatment with either RSK1 inhibitor or RSK inhibitor, blocking all isoforms of RSK, effectively reduced the COX-2 protein levels (Figure 2e). Collectively, the results from Figures 1 and 2 suggested that v-FLIP-mediated COX-2 transcription depends on NF-κB, CREB and MAPKs.
Role of v-FLIP/K13 transduction-induced MAPKs in COX-2 gene expression, protein levels and PGE2 secretion. (a) Phospho-MAPK array blots for the lysates prepared from p-SIN and v-FLIP/K13 HMVECs. (b) Densitometric analysis of MAPK array blots measuring the activation of MAPKs. The values were normalized to identical background levels using the R & D Systems (Minneapolis, MN, USA) Human MAPK antibody array analysis tool. The fold induction of MAPK was calculated by dividing the respective values obtained from v-FLIP/K13-HMVEC lysates with the values obtained from p-SIN-HMVEC lysates. (c) Cell lysates prepared from p-SIN and v-FLIP/K13-HMVECs were immunoblotted using p-ERK, p-AKT1, p-P65, p-RSK1 and p-GSK3-β antibodies. These blots were stripped and blotted against total antibodies for ERK, AKT, P65, RSK1, GSK3-β and tubulin to ensure equal protein loading. (d) v-FLIP/K13-HMVECs were treated with noncytotoxic concentrations of various MAPKs inhibitors for 1 h at 37°C. Total RNA was prepared from these cells and COX-2 gene expression was quantitated as described before. The COX-2 level in uninfected cells was considered as onefold for comparison. Percentage inhibition was calculated by considering COX-2 gene expression in inhibitor-untreated, v-FLIP/K13-transfected cells as 100%. (e) HMVEC-d cells were treated as described in (d). Total protein was prepared from these cells and COX-2/COX-1 protein levels were quantitated by western blotting. % Inhibition was calculated by considering the COX-2 protein level in inhibitor-untreated, v-FLIP/K13-transfected cells as 100%. β-Actin levels were checked as loading control. (f) Supernatants collected from the HMVECs treated in (d) were used for PGE2 measurement by ELISA as described before. Data are expressed as mean pg/ml. Each reaction was done in triplicate and each point represents the average±s.d. of five experiments. % Inhibition was calculated by considering PGE2 secretion from inhibitor-untreated KSHV-infected (2 h) cells as 100%. * and **: statistically significant at P<0.01 and P<0.005, respectively.
Consequence of inhibiting COX-2 in v-FLIP-dependent inflammation-related events
COX-2 inhibition modulates v-FLIP/K13 transduction-induced multiple chemokine ligands, chemokine-related genes and matrix-metalloproteinasess
Pretreatment of endothelial cells with chemical nonsteroidal anti-inflammatory drugs (NSAID), such as Indomethacin or COX-2-selective inhibitor NS-398 before KSHV infection, have been shown to abrogate PGE2 secretion in our previous study.5 Conventional NSAIDs have been shown to cause serious and significant complications.18 To evaluate the role of COX-2, we chose two COX-2-specific inhibitors, NS-398 and celecoxib. Though the selective COX-2 inhibitors only cause occasional deleterious effects, they have also been shown to exhibit some COX-2-independent effects.18, 19 Celecoxib, a selective COX-2 inhibitor (IC50=0.04 μM),20 was approved by the FDA in 1999.21 Celecoxib has a proven efficacy in modulating the promotion/progression stage of colon carcinogenesis,22 but its merits have never been explored in KSHV-associated malignancies and lymphomas expressing high levels of COX-2/PGE2.5, 6, 7, 9 Therefore, we first standardized the suitable non-cytotoxic dose of celecoxib in HMVEC-d cells using LDH release and a cell death ELISA (Supplementary Figures S3Aa and b). Similar to results obtained with the cell death ELISA, higher concentrations of celecoxib induced the cleavage of PARP in HMVECs (Supplementary Figure S3Ac). On the basis of data presented in Supplementary Figure S3A, we chose a 5 μM concentration of celecoxib throughout this study. We used a noncytoxic dose of NS-398 for the indicated times in this study, as we had thoroughly tested and described the use and effect of NS-398 on HMVECs in our previous studies.5, 6 We also found that v-FLIP-HMVEC cells were more sensitive to celecoxib and NS-398 treatment in comparison with p-SIN-HMVECs (Supplementary Figure S3B).
To understand the role of COX-2 in various v-FLIP/K13-induced inflammation-related genes, we prepared cDNA from serum-starved (8 h) cells (v-FLIP/K13-HMVECs) pretreated with either NS-398 (50 μM) or celecoxib (5 μM). Likewise, cDNA was prepared from serum-starved (8 h) p-SIN-HMVECs, and gene expression of chemokine ligands, chemokines, adhesion molecules and matrix-metalloproteinases (MMPs) were quantitated as indicated (Figure 3). We observed upregulated expression of all the genes tested in v-FLIP/K13-HMVECs when compared with p-SIN-HMVECs (Figures 3A–D). Pretreatment with NS-398 or celecoxib effectively reduced the expression of chemokine ‘C–X–C’ ligands (CXCL-2; GRO α/β, CXCL-5, CXCL-6; granulocyte chemotactic protein 2) (Figures 3Aa, c and d), but not CXCL-3 (GRO γ) (Figure 3Ab), while a stronger reduction was observed in (CXCL-5 and CXCL-6) (Figures 3Ac and d). Pretreatment with either NS-398 or celecoxib inhibited the expression of chemokines such as IL-8 (Figure 3Ba) and IL-6 (Figure 3Bb). The COX-2 inhibition reduced the expression of v-FLIP/K13-induced adhesion molecules ICAM-1 (Figure 3Bd), VCAM-1 (Figure 3Bc) and E-selectin (CD62 antigen-like family member E) (Figure 3Be). Strong reduction in ICAM-1 and VCAM-1 gene expression was observed in the celecoxib-treated v-FLIP/K13-HMVEC cells (Figures 3Bc and d). COX-2 inhibition reduced the expression of C–C ligand-related (CCL-2; MCP-1, CCL-5; RANTES-2, and CCL-20; MIP3α) genes (Figures 3Ca–c), which are especially important for the recruitment of a variety of immunoresponsive cells (monocytes, T lymphocytes and NK cells).
Effect of COX-2 inhibition on v-FLIP/K13-induced C–X–C ligands, C–C ligands, chemokines, adhesion molecules, MMP gene expression and MMP secretion levels. (A–C) Histograms depict the fold induction in gene expression of p-SIN, v-FLIP/K13-HMVECs, NS-398 or celecoxib-pretreated (4 h, 8 h and 24 h) v-FLIP/K13-HMVECs. Fold induction of chemokine (C-X-C motif) ligands (Aa, Ab, Ac, Ad), chemokine related genes (Ba, Bb, Bc, Bd, Be), chemokine (C-C motif) ligands (Ca, Cb, Cc) gene expression was calculated by considering expression in p-SIN-HMVECs as onefold. % Inhibition was calculated by considering gene expression in untreated v-FLIP/K13-HMVECs at the respective time of measurement as 100%. Each reaction was done in quadruplicate, and each bar represents the average±s.d. of four independent experiments. * and **: statistically significant at P<0.01 and P<0.005, respectively. (D) Effect of COX-2 inhibition on MMP secretion levels and invasion of v-FLIP/K13-HMVECs; activation of MMP-2, MMP-9 and MMP-10 measured by their respective assay kits in conditioned media obtained from untreated p-SIN-HMVECs and untreated, NS-398 or celecoxib-pretreated v-FLIP/K13-HMVECs. The levels of total and active MMPs in p-SIN-HMVECs were considered onefold for comparison. % Inhibition in these protease levels (total or active) upon COX-2 inhibition was calculated considering respective MMP levels (total or active) in untreated v-FLIP/K13-HMVECs as 100%. (E) COX-2 regulates v-FLIP/K13-HMVECs invasion via autocrine and paracrine mechanisms. Invasion of HMVEC cells in the presence of supernatant from pSIN or v-FLIP/K13-transduced or NS-398- or celecoxib-pretreated v-FLIP/K13-HMVECs. (a) Histogram represents the fluorescence intensity (mean±s.d. of three independent experiments) of HMVEC-d cells invaded in the presence of conditioned media obtained from pSIN or v-FLIP/K13 or NS-398/ celecoxib/ solvent control pretreated v-FLIP/K13-HMVECs. % Inhibition in invasion upon either NS-398 or celecoxib treatment was calculated considering invasion in the presence of supernatant from v-FLIP/K13-HMVECs at the indicated time point as 100%. (b) Effect of v-FLIP/K13-induced COX-2-mediated invasive potential of v-FLIP/K13-HMVECs as measured by fluorescence-based invasion assay as described in Materials and methods. Fluorescence intensity is presented and the values shown are the mean±s.d. of three independent experiments. % Inhibition in invasion by NS-398 or celecoxib treatment was calculated considering invasion of untreated v-FLIP/K13-HMVECs as 100%. Fold increase in the invasion of v-FLIP/K13-HMVECs was calculated using the invasive potential of p-SIN-HMVECs as onefold. Fluorescence associated with the invaded cells is shown and the values correspond to the mean±s.d. of three independent experiments. * and **: statistically significant at P<0.01 and P<0.005, respectively.
COX-2 inhibition regulates v-FLIP/K13-induced MMP gene expression, release of invasive factors and invasion of HMVEC-d cells
v-FLIP/K13 transduction strongly upregulated the expression of MMP-10, when compared with MMP-2 and MMP-9 (Figures 3Da–c). Pretreatment with COX-2 inhibitors specifically downregulated the expression of MMP-10 (Figures 3Da–c), when compared with MMP-2 and MMP-9 expression. A similar effect was observed in the secretion pattern of total and active MMPs (Figures 3Dd–f). To assess the functionality of active MMP secretion upon v-FLIP/K13 transduction, we performed invasion assays as described in the methods section. Figure 3Ea represents the invasive potential of the supernatants as analyzed by a Chemicon cell invasion assay and Figure 3Eb represents the data obtained using the Innocyte cell invasion assay depicting the intrinsic invasion of v-FLIP/K13-HMVECs. Without chemoattractant gradients, the intrinsic invasiveness of v-FLIP/K13-HMVECs through an ECM barrier was higher when compared with p-SIN-HMVECs (Figure 3Ea), suggesting v-FLIP is involved in invasion. This intrinsic invasion was reduced by 75% upon treatment with either NS-398 or celecoxib. To demonstrate whether COX-2 induction is important in promoting v-FLIP/K13-mediated cell invasion in a paracrine manner, we assessed the invasiveness of normal HMVEC-d cells in the presence of supernatants from untreated, NS-398, or celecoxib-treated v-FLIP/K13-HMVEC cells (Figure 3Ea), and observed a significant reduction in paracrine invasion as indicated (Figure 3Ea). Collectively, the results shown in Figure 3 indicate that v-FLIP/K13-induced COX-2 has critical roles in the regulation of MMP-10 and its associated invasion via autocrine and paracrine mechanisms.
Consequences of inhibiting COX-2 in v-FLIP-dependent tumorigenic events
COX-2 inhibition induces ‘anoikis’ as a novel mechanism of its antitumor effect
ECs die by apoptosis when detached from the ECM23 through a phenomenon called ‘anoikis’. Anoikis functions as a surveillance mechanism to preserve normal tissue architecture by eliminating the cells attempting to deviate from their normal spatial constraints.24 As cells become transformed, they gain the ability to grow in the absence of contacts with a solid ECM and attain the additional capacity for invasion and metastasis, while acquiring resistance to anoikis. Inhibition of anoikis is a critical step in tumor progression and metastatic spread because it enables malignant cells to detach from the primary tumor and invade the secondary tissues. Therefore, new therapies directed to sensitize tumor cells to ‘anoikis’ are especially relevant as they could have both antitumor and antimetastatic properties. Since celecoxib and NS-398 have been shown to induce anoikis in many cancer cell types16, 25 and v-FLIP has been previously shown to resist anchorage-independent cell death,15 we hypothesized that KSHV v-FLIP utilizes host molecules such as COX-2/PGE2 to evade anoikis. We addressed this question in three parts: (1) understanding the effect of detachment-induced changes in the formation of focal adhesions (FAs) and the effect on ECM interaction-induced cell signaling cascades at very early points of detachment (2 h); (2) consequences of COX-2 inhibition on the cell survival of v-FLIP-transduced and normal ECs in suspension (12 h); and (3) the effect of COX-2 inhibition on colony formation (6 days).
COX-2 inhibition has a role in NF-κB activity and affects changes in the detachment-induced cytoskeletal rearrangement events in v-FLIP/K13-HMVEC-d cells at early time points
In an inactive state, NF-κB is located in the cytoplasm as a heterotrimer consisting of p50, p65 and the inhibitory subunit of NF-κB (IαB) α subunits. In response to an activation signal, the IκBα subunit is phosphorylated, ubiquitinated and degraded through the proteasomal pathway, thus exposing the nuclear localization signals on the p50–p65 heterodimer. The p65 subunit is then phosphorylated, leading to nuclear translocation and binding to a specific DNA sequence, which in turn results in gene transcription. An IκBα kinase, IKK, has been identified in phosphorylating serine residues in IκBα at positions 32 and 36.26 Untreated v-FLIP/K13-HMVEC cells displayed intense p65 nuclear staining (Figure 4Aa (1 and 2)), whereas NS-398 or celecoxib treatment effectively reduced the nuclear translocation of p65 (Figure 4Aa (3–6)).
(A) Effect of COX-2 inhibition on detachment-induced cytoskeletal rearrangement-linked cellular survival kinases. (a) p65 immunostaining in untreated, NS-398- or celecoxib-treated v-FLIP-HMVECs. (b) EMSA for nuclear extracts prepared from untreated, NS-398- or celecoxib-treated v-FLIP-HMVECs using the NF-κB probe. This EMSA experiment shows the binding of nuclear extracts prepared from untreated, NS-398- or celecoxib-treated v-FLIP-HMVECs to the NF-κB probe. The specificity of the DNA–protein interaction was assessed by competition using a 100-fold molar excess of unlabeled double-stranded oligonucleotide NF-κB probe as a competitor. Each EMSA is representative of at least three independent experiments. (c) Cell lysates prepared from untreated, NS-398- or celecoxib-treated v-FLIP-HMVECs were western blotted using p-FAK, p-Src, p-AKT, p-P65, p-ERK and laminin-γ antibodies. These blots were stripped and blotted against total antibodies for FAK, Src, AKT, P65, ERK and ß-actin to confirm equal protein loading. (Ba) Representative pictures depicting p-FAK, p-Src, p-FAK/p-Src and focal adhesions. (b) Table representing the number of cells with p-FAK, p-Src or p-FAK/p-Src staining per 10 cells. (C) Laminin-γ staining of untreated or NS-398-treated v-FLIP-HMVECs by immunofluoresence microscopy. White arrows in B and C denote FAK-Y397, p-Src, FAK-Y397/p-Src and Laminin-gamma. (D) Histogram representing Rac1-GTPase activation as measured by G-LISA. Fold activation of Rac1 is calculated by considering Rac1-GTPase activity in the p-SIN-HMVECs as onefold. Each reaction was done in triplicate, and each bar represents the mean±s.d. for three experiments. *statistically significant at P<0.01.
To determine the effect of celecoxib/NS-398 treatment on NF-κB activation, we carried out electrophoretic mobility shift assay (EMSA). Briefly, nuclear extracts prepared from untreated or COX-2 inhibitor-pretreated v-FLIP/K13-HMVECs were incubated with 32P end-labeled double-stranded NF-κB oligonucleotide (Figure 4Ab; lanes 1–6). Nuclear extracts prepared from untreated v-FLIP/K13-HMVECs (Figure 4Ab; lanes 1 and 4) showed complex formation, whereas nuclear extracts prepared from celecoxib/NS-398-treated (Figure 4Ab, lane 2) cells showed reduced complex formation (Figure 4Ab; lanes 2 and 5). A competition assay with an excess of unlabeled cold probe (NF-κB) completely abolished complex formation with labeled probe, confirming the specificity of the assay and validating that the complex is specific to NF-κB (Figure 4Ab; lanes 3 and 6). These findings suggest that celecoxib and NS-398 suppress NF-κB activation, which might explain their antitumor, chemopreventive, antiangiogenesis, antiproliferative and apoptosis-inducing activities. This data was also confirmed by NF-κB ELISA (Supplementary Figure S4A). Celecoxib- and NS-398-treated v-FLIP/K13-HMVEC cells showed 77% and 87% inhibition of p65 activation, respectively, when compared with untreated cells. Solvent treatment did not lower p65 activation (Supplementary Figure S4A) reaffirming the role of COX-2 inhibition in NF-κB activity downregulation.
Though the mechanisms of anoikis are not completely understood yet, recent reports suggest that deprivation of integrin-mediated survival signals is one of the major causes of anoikis. Cellular adhesion to the matrix is mainly mediated by integrins, which provide survival signals through the activation of the PI-3K/AKT, Ras/Raf/ERK or JNK pathways. Among these, AKT seems to be a central molecule in cellular survival signaling because FAK and ILK, both of which are integrin-associated non-receptor kinases, converge at the activation of AKT. AKT executes its survival function, in part, by inhibition of caspase-9 and Bad. Focal contacts/FAs are dynamic structures of close interaction between cells and the matrix on their substrate.27 These contacts are dynamic structures that show motility even in stationary cells and are the conduits of signals from the matrix to the cytoplasm of the cell and vice versa.28 The extracellular domains of integrins bind matrix elements such as fibronectin, vitronectin and laminin in the connective tissue, thereby providing a structural link between the ECM and the actin-based cytoskeleton system of a cell.29 Molecular composition of FAs includes proteins such as vinculin, paxillin and tensin, but these are also described as hubs for survival signals and have been shown to be critical for cell survival. FAs are responsible for the activation of AKT, ERK and NF-κB. COX-2 inhibition could downregulate the phosphorylation of FAK, Src, AKT and p65, although there was no effect on ERK phosphorylation (Figure 4Ac). We also checked for another molecule called laminin-γ1, whose gene expression was found to be upregulated (eightfold) upon v-FLIP transduction in ECs (data not shown; Figure 4Ac). Laminins, a family of ECM glycoproteins, are the major noncollagenous constituent of basement membranes. They have been implicated in a wide variety of biological processes including cell adhesion, differentiation, migration, signaling and metastasis. We reasoned that increased laminin-γ1 might be supporting faster attachment, spreading and enhancing cell to cell connection/cell survival in v-FLIP-transduced ECs, all the features expected for KS cells with a spindle phenotype. COX-2 inhibition in v-FLIP/K13-HMVEC cells effectively reduced the levels of laminin-γ1 (Figure 4Ac).
As the protein phosphorylation data were encouraging, we assessed a few important events at the FAs that could push the cells toward improved and faster cell survival, such as the number of FAs, size of FAs, number of FAs with FAK or Src phosphorylation, and the number of FAs with FAK/Src colocalization (Figure 4B). Immunofluorescence was conducted and the images presented are high-resolution deconvoluted images with no nearest neighbors. Untreated v-FLIP-HMVECs were strongly stained for FAK (Tyrosine 397) and Src (Tyrosine 416) phosphorylation (Figure 4Ba (1–12)). We also observed reasonably large and well-defined FAs (Figure 4Ba (3 and 9)). We counted an average of 240 points of focal contacts per 10 cells (Figure 4Bb). Pretreatment with COX-2 inhibitors reduced the number of FAs by 61% (Figure 4Bb). We observed that v-FLIP-HMVECs could attain the elongated phenotype faster, whereas COX-2 inhibitor-treated cells appeared rounded and smaller when compared with untreated cells (Figure 4Bb (4, 10, 13 and 14)). Colocalization of the phosphorylated forms of FAK and Src at FAs had previously been shown to affect the downstream signaling events responsible for the survival of ECs in other systems; therefore, we checked p-FAK and p-Src colocalization (Figure 4Bb (6 and 12)). Pretreatment with COX-2 inhibitor reduced the number of FAs showing colocalization of p-FAK and p-Src (Figure 4Bb (16)) when compared with untreated cells (Figure 4Bb (6 and 12)). We also observed laminin-γ1 staining in the stress fiber-like projections emanating from the v-FLIP/K13-HMVECs (Figure 4C (1–7)). Untreated cells had a higher number of these protrusions and had a higher number of cell/cell connections as signatures of good cell spreading and cell/cell communication (Figure 4C (1, 2, 3 and 7)). COX-2 inhibitor-pretreated cells had smooth cell surface edges and lesser cell-to-cell connections (Figure 4C (4–6).
COX-2 inhibition downregulates Rac-1 activation in v-FLIP-HMVECs
Rearrangement of the actin cytoskeleton is primarily controlled by members of the Rho-GTPase family such as RhoA, Rac-1 and Cdc42.30 Our earlier studies have demonstrated the activation of these GTPases by KSHV infection, interaction of the KSHV envelope glycoprotein gB with adherent ECs or fibroblast cell integrins and in the presence of PGE2 released from infected ECs.6, 31 Rac1-GTPases have not only been shown to be critical regulators of vesicle trafficking, gene transcription, cell adhesion and cell spreading but have also been shown to be responsible for cell transformation, anchorage-independent cell growth and anoikis resistance32 via NF-κB activation. We hypothesized that COX-2-induced PGE2 might be involved in providing anoikis resistance to v-FLIP/K13-HMVECs via the upregulation of Rac1-GTPase activation. We assessed the role of COX-2 inhibition on the activation kinetics of Rac1-GTPase by Rac1-GLISA (Figure 4D). We observed a 4.4-fold induction of Rac1-GTPase in v-FLIP/K13-HMVECs when compared with p-SIN-HMVECs (Figure 4D). Pretreatment with NS-398 or celecoxib effectively reduced the levels of Rac1-GTPases in v-FLIP-HMVEC-d cells, by 52% and 59%, respectively (Figure 4D). We observed a 3.8-fold induction of RhoA-GTPase in v-FLIP/K13-HMVECs when compared with p-SIN-HMVECs (Supplementary Figure S4B). Pretreatment with NS-398 or celecoxib could not reduce the levels of RhoA-GTPase in v-FLIP-HMVEC cells (Supplementary Figure S4B).
v-FLIP/K13 induces signaling molecules (FAK, Src, AKT, ERK, P-38, RSK and Rac1) indirectly through secretion of cytokines
Our study presented multiple evidences for v-FLIP-mediated direct activation of signal pathways (Figures 2A, B, C and 4C). In order to study the role of cytokines secreted from v-FLIP-transduced cells in signal cascade induction, we collected supernatants from serum-starved (8 h) p-SIN and v-FLIP/K13-HMVEC cells (Figure 5A). v-FLIP-HMVEC cells used for collecting supernatants had characteristic elongated spindle phenotype. These supernatants were used to induce signaling in serum-starved (8 h) HMVEC cells for 4 h, 8 h and 24 h. Lysates prepared from these cells showed activation for all the signal molecules (FAK, Src, AKT, ERK, P-38 and RSK) tested though the time point of their induction varied as indicated (Figure 5b). Cells incubated with cytokines from v-FLIP-transduced cells showed higher phosphorylation of FAK and P-38 at 4 h (Figure 5b) when compared with p-SIN. Phosphorylation of Src and ERK in cells induced with cytokines from v-FLIP/K13-HMVECs was higher compared with that observed from p-SIN cells at all the time points tested (Figure 5b). We observed increased phosphorylation of AKT at 8 h and 24 h in the cells induced with the supernatant obtained from v-FLIP/K13-HMVEC cells (Figure 5b). Higher RSK activation was observed in the cells stimulated with the supernatant obtained from v-FLIP/K13-HMVEC cells for 4 h and 8 h when compared with the levels in the cells induced with the supernatant obtained from p-SIN-transduced cells (Figure 5b).
Indirect role of v-FLIP-induced cytokines in the induction of various signaling molecules. (a) Cell lysates prepared from serum-starved (8 h) HMVECs incubated with supernatants collected from serum-starved (8 h) p-SIN and v-FLIP/K13-HMVECs cells for 4 h, 8 h and 24 h. (b) Cell lysates were western blotted using p-FAK, p-Src, p-AKT, p-P38, p-ERK and p-RSK antibodies. These blots were stripped and blotted with total antibodies against FAK, Src, AKT, P38, ERK and RSK. (c) Cell lysates prepared from serum-starved (8 h) HMVECs incubated with supernatants collected from untreated serum-starved (8 h) p-SIN, untreated v-FLIP/K13-HMVECs and v-FLIP/K13-HMVECs pretreated with either NS-398 or celecoxib for 8 h. These supernatants were used to induce signaling in serum-starved (8 h) HMVECs for 4 h for FAK, Src, ERK, P-38 and RSK, and 8 h for AKT. (d, e) Rac1-GTPase activity in the lysates prepared in (b) and (c). Histogram representing Rac1-GTPase activation as measured by G-LISA. Fold activation of Rac1 is calculated by considering Rac1-GTPase activity in the p-SIN-HMVECs as onefold. Each reaction was done in triplicate, and each bar represents the mean±s.d. for three experiments. * and **: statistically significant at P<0.01 and P<0.005, respectively. The % Inhibition in invasion by NS-398 or celecoxib treatment was calculated considering invasion of untreated v-FLIP/K13-HMVECs as 100%.
Since our previous results showed effective downregulation of cytokine gene expression (Figure 3) in the cells pretreated with COX-2 inhibitors, we reasoned that COX-2 inhibition might change the level of cytokine secretion also. We collected supernatants from serum-starved (8 h) p-SIN, v-FLIP/K13-HMVECs and v-FLIP/K13-HMVECs pretreated with either NS-398 or celecoxib for 8 h. These supernatants were used to induce serum-starved (8 h) HMVECs for 4 h to test the activation of FAK, Src, ERK, P-38 and RSK, and 8 h for AKT (higher activity of AKT was observed at 8h; Figure 5b). We observed effective downregulation in the activation of all signal molecules tested in the lysates prepared from the cells incubated with supernatants obtained from COX-2 inhibitor-treated cells when compared with the supernatants collected from untreated cells (Figure 5c).
Similarly, we measured the activity of Rac1-GTPase in the lysates used in Figures 5b and 5c. Activity of Rac1-GTPase in cells induced with cytokines from v-FLIP/K13-HMVECs was higher when compared with the activity observed from p-SIN cells at all the time points tested (Figure 5d). We observed downregulation in the Rac1-GTPase activity in the lysates prepared from the cells incubated with supernatants obtained from COX-2 inhibitor-treated v-FLIP/K13-HMVEC cells when compared with the supernatants collected from untreated cells (Figure 5e). Our results convincingly show the interplay of indirect activation of signaling pathways mediated via cytokines secreted and spindle cell differentiation induced by the expression of KSHV oncoprotein v-FLIP.
COX-2 inhibition downregulates MnSOD level in v-FLIP-HMVECs
MnSOD has been shown to have roles in cancer cell growth, metastasis promotion and protection from tumor surveillance-mediated apoptotic mechanisms. We hypothesized that COX-2-induced PGE2 might be involved in providing antioxidant support to enhance anoikis resistance in v-FLIP/K13-HMVECs. We assessed the role of COX-2 inhibition on antioxidant status by measuring superoxide dismutase activity (Supplementary Figure S4C) as described in Materials and methods. We observed a 4.4-fold induction of MnSOD in v-FLIP/K13-HMVECs when compared with p-SIN-HMVECs (Supplementary Figure S4C). Pretreatment with NS-398 or celecoxib effectively reduced the levels of MnSOD in v-FLIP-HMVEC-s, by 84% and 55%, respectively (Supplementary Figure S4C).
COX-2 inhibition abrogates v-FLIP-mediated protection of HMVECs from anoikis
It is well documented that COX-2 expression correlates with tumor development and progression in most human tumors. COX-2 has been known to act as a survival factor under a variety of cellular stress conditions. We examined the effect of COX-2 inhibition (NS-398 or celecoxib pretreatment) on anoikis resistance-mediated cell survival in v-FLIP-HMVECs (Figure 6). Anoikis propelled cell death and changes in cell viability were monitored microscopically for the presence of red EthD-1 fluorescence (Figure 6Af and g) and green Calcein AM, respectively (Figure 6Ab, c, d and e).
(A) Effect of COX-2 inhibition on v-FLIP-mediated anoikis resistance. (a) Schematic for anoikis assay. (b) Green Calcein AM staining in attached untreated, solvents for either celecoxib or NS-398-treated, celecoxib or NS-398-treated p-SIN and v-FLIP-HMVECs. (c) Green Calcein AM staining in suspended untreated, solvents for either celecoxib or NS-398-treated, celecoxib or NS-398-treated p-SIN and v-FLIP-HMVECs. (d) Green fluorescence units measured as index of viability in the lysates prepared from the attached and suspended cells shown in (b, c). (e) MTT assay readings at 570 nm measured as index of proliferation in attached and suspended cells shown in (b, c). MTT cell proliferation assay measures cell metabolism and is based on the cleavage of the yellow tetrazolium salt MTT to purple formazan crystal by metabolically active cells. (f) Ethidium homodimer staining in suspended untreated, solvents for either celecoxib or NS-398-treated, celecoxib or NS-398-treated p-SIN and v-FLIP-HMVECs. EthD-1 penetrates damaged cell membranes, fluoresces with a 40-fold enhancement upon binding ssDNA, dsDNA, RNA, oligonucleotides and triplex DNA. (g) Red fluorescence units measured as index of cell death in the lysates prepared from suspended cells. (Ba) MTT assay readings at 570 nm for p-SIN and v-FLIP-HMVECs suspended in the presence of supernatant from either p-SIN or v-FLIP-HMVECs depicting the significance of paracrine survival. (b) MTT assay readings at 570 nm for v-FLIP-HMVECs cultured in the presence of untreated, solvents for either celecoxib or NS-398-treated, or celecoxib or NS-398-treated v-FLIP-HMVECs, depicting the significance of paracrine survival. Red arrows pointing down denote decrease.
The anoikis assay was performed as described in the Materials and methods section and in Figure 6Aa. In adherent cultures of untreated/celecoxib or NS-398 solvent-treated and celecoxib/NS-398-treated v-FLIP or p-SIN-HMVEC cells, the basal level for apoptosis was negligible, and cell viability (as shown by Calcein AM staining) was 100% (Figure 6Ab (1–10)). Anoikis was induced after 12 h of detachment on an anchorage-resistant plate, and p-SIN-HMVECs entered apoptosis and had reduced viability as indicated by lower staining of suspended cells by Calcein AM (Figure 6Ac (6)). However, cells expressing v-FLIP were highly resistant to anoikis, with an increased number of viable cells after detachment (Figure 6Ac (1)). Cell viability was also measured using the Calcein AM (485 nm/515 nm) fluorimetric detection method that showed results similar to our microscopy observations (Figure 6Ad). Similarly, we tested the proliferation index in cells undergoing anoikis using an 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT) assay (Figure 6Ae). MTT assay results matched cell viability and microscopic analysis results (Figures 6Ab–e) further validating our hypothesis.
Anoikis propelled cell death was measured by EthD-1, an excellent marker for measuring dead cells. As shown in Figures 6Af and g, p-SIN-HMVECs showed increased cell death upon plating on anchorage-resistant plates (Figures 6Af and g), further suggesting that anoikis resistance is provided by v-FLIP. Untreated v-FLIP/K13-HMVECs exhibited reduced cell death as compared with NS-398 or celecoxib-pretreated cells as indicated (Figures 6Af and g), whereas solvent-pretreated v-FLIP/K13-HMVECs did not show any increase in cell death; levels were similar to untreated cells confirming drug specificity (Figures 6Af and g).
Our results presented in Figure 3 clearly demonstrated that COX-2 inhibition by either NS-398 or celecoxib could reduce gene expression as well as secretion of inflammation/invasion/growth-related factors. We therefore speculated that the reduced secretion of growth and survival factors from COX-2 inhibitor-pretreated v-FLIP/K13-HMVECs might affect the viability of neighboring untransduced cells undergoing anoikis-associated cell death via paracrine pathways. We assessed the viability of p-SIN-HMVECs in the presence of supernatants obtained from either p-SIN or v-FLIP-HMVECs (Figure 6Ba). Supernatants obtained from COX-2 inhibitor-treated cells could effectively reduce anoikis resistance through a paracrine mode, probably via decreased secretion of growth factors (Figure 6Bb). Our results (Figure 6Bb) suggested that expression of v-FLIP not only protects cells that are positive for v-FLIP expression but can also rescue neighboring cells from anoikis.
Along with the data shown in Figure 4Aa, it is possible that COX-2/PGE2 help in the maintenance of NF-κB signaling in HMVEC cells that is responsible for sustained COX-2 levels, and eventually this loop of events helps maintain intrinsic survival signals, and continuous secretion of inflammation, invasion, angiogenesis and growth-related factors. ECM induced the NF-κB, FAK/Src, PI3K-AKT pathways, along with secreted growth factors such as VEGF-C, chemokines such as IL-8, MCP-1 and IL-6, and invasive factors such as MMP-10 thereby enhancing v-FLIP-mediated anoikis resistance.
COX-2 inhibition results in the decreased transformation potential of v-FLIP as measured by a colony formation assay
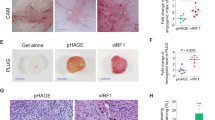
v-FLIP/K13-HMVECs are known to undergo neoplastic transformations that yield a cell population capable of proliferating independently of both external and internal signals. As v-FLIP/K13-HMVECs showed reduced requirements for extracellular growth-promoting factors, were not restricted by cell-cell contact and showed anchorage-independent growth phenomenon, we wanted to understand the effect of COX-2 inhibition on v-FLIP/K13-mediated transformation (Figure 7). Traditionally, the soft agar colony formation assay is a common method to monitor anchorage-independent growth, which measures proliferation in a semisolid culture media after 3–4 weeks by manual counting of colonies, however, we assessed the effect of COX-2 inhibitor treatment on the transformation efficiency of v-FLIP/K13 ECs by a modified cell transformation assay from Cell Biolabs as described in Materials and methods and in Figure 7Aa. v-FLIP/K13-HMVECs pretreated with solvent control or cells pretreated with COX-2 inhibitors or supplemented with either solvent control or COX-2 inhibitors were utilized for transformation assays. This assay is quick as it does not require a 3–4 week incubation period and is highly reliable as it does not involve manual counting of colonies, which could be highly subjective. Instead, cells are incubated only 6 days in a proprietary semisolid agar media before being solubilized, transferred and detected by MTT (Figure 7Ac), which is highly sensitive and quantitative. Changes in the number of colonies formed were captured by microscopy (Figure 7Ab). The crystal violet (blue) stain in the colonies reflects ‘live’ colonies (Figure 7Ab). To be more precise with quantitation of colonies, we solubilized one set of colonies and stained these for their metabolic activity using MTT (Figure 7Ac). Untreated v-FLIP-HMVECs formed a reasonably large number of colonies, whereas p-SIN-treated HMVECs did not (data not shown). COX-2 inhibitor pretreatment followed by replacing the growth medium of v-FLIP-HMVECs with COX-2 inhibitor drastically reduced the formation of colonies when compared with untreated cells (Figure 6Ab (1, 3 and 5)). Pretreatment with COX-2 inhibitors such as celecoxib and NS-398 reduced the colony formation by 57% and 72%, respectively, as determined by MTT assay (Figure 7Ac). Pretreatment with solvent for reconstituting celecoxib or NS-398 had no effect on colony formation, thus validating the specificity of COX-2 inhibitor treatment. The colony numbers were similar to the ones formed by untreated cells (Figure 7Ab (1, 2 and 4)).
(A) Effect of COX-2 inhibition on v-FLIP/K13-induced colony formation and EndMT-linked genes. (a) Transformation assay schematic and recovery of transformed cells. (b) Untreated or COX-2 inhibitor-treated or solvent control-treated v-FLIP-HMVECs were plated in soft agar in the presence of the indicated doses of drugs or solvent controls at indicated times, and colonies were scored after 6 days. The values shown are mean±s.d. of two independent experiments performed in duplicate. Crystal violet staining of colonies formed from untreated, celecoxib or NS-398 solvent-treated and celecoxib or NS-398-treated v-FLIP-HMVECs. Light microscopy fields (original magnification, × 40) of untreated or COX-2 inhibitor-treated or solvent control-treated v-FLIP-HMVECs are shown. (c) MTT assay for the colonies solubilized from untreated, celecoxib or NS-398 solvent-treated and celecoxib or NS-398-treated v-FLIP-HMVECs. Histogram showing the MTT for the colonies shown in panels. The values shown are mean±s.d. of two independent experiments performed in duplicate. (B) Expression of (a) snail, (b) slug, (c) twist, (d) laminin-γ, (e) E-cadherin in the cells obtained from colonies derived in (A) * and **: statistically significant at P<0.01 and P<0.005, respectively.
As colony formation may also be affecting the phenotype of ECs and executing some EndMT, we assessed a few genes exclusive for this phenomenon in v-FLIP-HMVECs. EndMT is a phenomenon where little experimental advancement has been made, and the putative mediators of EndMT include factors such as Wnt, TGF-β, Jagged/Notch, PAR-1, FGF-2, HGF, RTK, integrins and MMPs. During EndMT, various phenotypic markers are lost (VE-cadherin, Tie-1/2, VEGFR-1/2, PECAM/CD31, and FVIII) and new markers are gained (α-Smooth muscle actin, type I and III collagens, vimentin and calponin). Dermal microvascular ECs, for example, are described as very susceptible to transitioning into mesenchymal cells.33 Keeping this in mind, we recovered the viable cells from the colonies of untreated or COX-2 inhibitor-treated v-FLIP-HMVECs and prepared cDNA. We tested for some of the EndMT-related genes such as slug, snail, twist, laminin-γ and E-cadherin as indicated (Figure 7B). Among these, ‘slug’ is a transcriptional repressor (a novel Notch target) and has been shown to be essential for Notch-mediated repression of E-cadherin, resulting in ß-catenin activation and resistance to anoikis.34 Slug's roles in endocardial cushion formation and early events of EC transformation have been established.35 Snail's expression has been linked to tumors with deeper invasive activity and high metastatic potential. Snail has also been shown to enhance the migration and proliferation activity of cancer cells. The ‘twist’ protein is a highly conserved transcription factor that has key roles in embryonic development and is frequently reactivated in a wide array of human cancers (sarcomas, gliomas, neuroblastomas and melanomas), where it invariably correlates with more aggressive, invasive and metastatic lesions. ‘Twist’, a potent inhibitor of the cell-safety program, displays abnormal mitogenic, antiapoptotic and metastatic properties, and its expression in tumor cells induces a widespread chemoresistance to therapeutic drugs (paclitaxel, daunorubicin, etoposide, doxorubicin and cisplatin) in various cancer types. Functional studies have indicated that ‘Twist’ may have a major role in tumor promotion and progression, by inhibiting differentiation, interfering with the p53 tumor-suppressor pathway and favoring cell survival, and/or inducing EndMT. Cadherins are comprised of a family of Ca+2-dependent adhesion molecules that function to mediate cell–cell binding critical for the maintenance of tissue structure and morphogenesis. The classical cadherins, E-, N- and p-cadherin, consist of large extracellular domains that are thought to be responsible for binding specificity, transmembrane domains and carboxy terminal intracellular domains. COX-2 inhibitor pretreatment and supplementation after 2 days during colony formation reduced the expression of slug, snail, twist and laminin-γ but enhanced the expression of E-cadherin (Figure 7A), highlighting that v-FLIP induced the colony formation and that EndMT in ECs can be efficiently downregulated by COX-2 inhibitors.
Mechanism of inhibiting COX-2 in v-FLIP-dependent growth
COX-2 inhibition reduced v-FLIP's transformation potential via ‘extrinsic’ and ‘intrinsic’ death receptor pathways
There are two major pathways of caspase activation, the extrinsic pathway, triggered by ‘death receptor’-ligand binding (FASL to FAS), and the intrinsic pathway, triggered by cellular stress, such as occurs after cytokine deprivation, DNA damage and anoikis. A critical feature of the intrinsic pathway includes the interplay of Bcl-2 family member proteins. Overexpression of some antiapoptotic family members, such as Bcl-2, BCL-xL, and stability of the Bcl-2-related antiapoptotic myeloid cell leukemia sequence 1 (MCL1) protein can prevent apoptosis induced by the withdrawal of GFs. Bcl-2-associated X protein on the other hand is a proapoptotic Bcl-2 family member. BAX translocates to the mitochondria during apoptosis, causing release of cytochrome c into the cytosol. A third proapoptotic subgroup of the Bcl-2 family includes BIM and BAD, which contain a ‘BH3’ protein domain that promotes apoptosis when overexpressed. In this study, we sought to determine the mode of cell death induced by anoikis in v-FLIP/K13-HMVECs. We anticipate that an improved understanding of this process could provide insight into the mechanisms of sensitivity and/or resistance to caspase inhibitors, shed light on the chemotherapeutic potential of celecoxib and NS-398, and uncover new targets for therapy.
The cellular inhibitor of apoptosis 1 and 2 (cIAP1 and cIAP2) proteins have been implicated in the activation of NF-κB and have been shown to stop apoptotic cell death by overproduction of caspases (3, 7, and 9). We hypothesized that IAPs may be mediating this resistance so knockdown of cIAP-1, cIAP-2 and the x-linked inhibitor of apoptosis protein (x-IAP) would increase sensitivity to apoptosis. To determine which Bcl-2 family members are potentially involved in the maintenance of v-FLIP-HMVECs, we studied the effect of celecoxib or NS-398 treatment on the expression of Bcl-2, BCL-xL, MCL1, BAX and BIM in v-FLIP-HMVECs. Both drugs (celecoxib and NS-398) reduced cIAP2 and x-IAP expression effectively when compared with cIAP1 (Figure 8A). Antioxidants, such as SOD, mitochondrial Mn-SOD, Cu/Zn-SOD and catalase are able to degrade hydrogen peroxide (H2O2), an important ROS related to p53 activation and protect a variety of cells against oxidative stress-induced toxicity.36 Celecoxib or NS-398 reduced the SOD2 levels as compared with the levels of SOD1 and catalase (Figure 8A). IEX-1 (p22/PRG1) is an early response NF-κB target gene implicated in the regulation of cellular viability, cell cycle progression and proliferation. Its overexpression can suppress or enhance apoptosis depending on the nature of the stress. Expression of BCL2A oncoprotein has been shown to inhibit programmed cell death (apoptosis). Celecoxib and NS-398 treatment effectively downregulated BCL2A and IEX-1 gene expression (Figure 8A).
Effect of COX-2 inhibition on the cell survival mechanism of v-FLIP/K13-induced anoikis resistance. (A) Expression of (a) cIAP-1, (b) cIAP-2, (c) xIAP, (d) catalase, (e) SOD1, (f) SOD2, (g) BCL2A and (h) IEX-1 in the cells obtained from colonies of untreated, celecoxib or NS-398 solvent-treated and celecoxib or NS-398-treated v-FLIP-HMVECs. (B) Protein levels of Bcl-2, BAX, PARP, cleaved PARP, MCL-1, BCL-xL, BIM and β-actin in the cell lysates prepared from colonies of untreated, celecoxib or NS-398 solvent-treated and celecoxib or NS-398-treated v-FLIP-HMVECs. Blots are representative of three independent experiments. (Ca, b) Celecoxib or NS-398 treatment induces colocalization of BAX and cytochrome c in v-FLIP- HMVECs undergoing anoikis. Untreated and COX-2 inhibitor-treated cells were fixed and immunostained with antibodies against BAX (red) and cytochrome c (green) or with DAPI to detect DNA (blue). Cells were imaged by confocal microscopy. (Da, b) Celecoxib or NS-398 treatment modulates levels of DR4 and DR5 at the protein level. Treated and untreated v-FLIP-HMVECs were stained using DR4 or DR5 antibody. Flow cytometry was performed with a FACS calibur flow cytometer and analyzed with CellQuest Pro software (San Jose, CA, USA). * and **: statistically significant at P<0.01 and P<0.005, respectively.
To determine which Bcl-2 family members are potentially involved in the maintenance of v-FLIP-HMVEC cells, we studied the effect of celecoxib or NS-398 treatment on the expression of Bcl-2, BCL-xL, MCL1, BAX and BIM in v-FLIP-HMVECs. We found that the expression levels of only the antiapoptotic protein MCL1 and the proapoptotic BH3-only protein BIM were significantly altered by either celecoxib or NS-398 treatment (Figure 8B). Next, we focused our attention on BIM, and did immunoblotting analysis of the untreated or celecoxib/NS-398-pretreated v-FLIP-HMVECs (Figure 8B). Results revealed that the levels of two major spliced isoforms-BIM-long (L) and BIM-short (S) were induced after celecoxib/NS-398 treatment in v-FLIP-HMVECs but not in untreated or solvent-treated cells (Figure 8B). Both the drugs (celecoxib and NS-398) induced apoptosis in v-FLIP-HMVECs, as indicated by the presence of PARP cleavage products in cell lysates (Figure 8B).
Change in the localization of proapoptotic BAX is another hallmark of the intrinsic pathway of apoptosis. Immunostaining followed by confocal microscopy revealed that at baseline (treatment with solvent for NS-398 or celecoxib) (Figures 8Ca (2) and Cb (2)) BAX was found in the nuclei of v-FLIP-HMVECs but following celecoxib (Figure 8Cb (5)) or NS-398 (Figure 8Ca (5)) treatment, BAX was found in the cytoplasm in apoptotic cells. The distribution of cytochrome c changed from punctate to more diffuse, consistent with the release of cytochrome c from the mitochondria. Merging of the two fluorescent signals (Figures 8Ca (6) and Cb (6)) revealed intense overlap between BAX and cytochrome c staining. These observations suggest that BAX translocates to the mitochondria during celecoxib- or NS-398-induced apoptosis in v-FLIP-HMVECs.
In order to find which caspase gets activated in the cells treated with COX-2 inhibitors, we observed the enhanced survival of cells incubated with or without COX-2 inhibitor in the presence of caspase-3 (executioner caspase), caspase 8 (initiator caspase) and caspase-9 (initiator caspase) inhibitors, suggesting the role of COX-2 inhibitors in inducing apoptosis (Supplementary Figure S5A). We also observed the activation of caspases 3, 8 and 9 in COX-2 inhibitor-treated suspended v-FLIP-HMVECs (Supplementary Figure S5B), which validated data in Supplementary Figure S5A. Members of the TNF receptor gene superfamily, such as the TNF-related apoptosis-inducing ligand (TRAIL) receptor, death receptor 4 (DR4; also called TRAIL-R1) and death receptor 5 (DR5; also named Apo2 or TRAIL-R2) share a similar, cysteine-rich extracellular domain and an additional cytoplasmic death domain. Both DR4 and DR5, located at the cell surface, become activated or trimerized on either binding to their ligand TRAIL or by overexpression and then signal apoptosis through caspase-8-mediated activation of caspase cascades. These DRs attract much more attention because their ligand TRAIL preferentially induces apoptosis in transformed or malignant cells, showing potential as a tumor-selective apoptosis-inducing cytokine for cancer treatment. The expression of DR4 and DR5 is inducible by certain stimuli, including some cancer therapeutic agents; NSAIDs have been shown to induce DR expression.37 Induction of DR4 and/or DR5 accounts for induction of apoptosis and/or enhancement of TRAIL-induced apoptosis by certain cancer therapeutic agents. Celecoxib or NS-398 treatment of v-FLIP-HMVECs enhanced the surface expression of DR4 (Figure 8Db) but not DR5 (Figure 8Db).
v-FLIP-induced COX-2 regulates growth of v-FLIP-HMVECs displaying anoikis resistance
As v-FLIP-induced COX-2 has as an important role in providing anoikis resistance, as seen by MTT assays, we assessed the effect of NS-398 or celecoxib on their cell cycle profile in cells undergoing anoikis. According to DNA profiling, a significantly higher proportion of untreated v-FLIP-HMVECs were in S-phase compared with either celecoxib or NS-398-treated cells. We observed a clear antiproliferative shift in cell cycle parameters towards a reduced percentage of cells at the S and G2/M phases, together with an increased percentage of cells at the G1 phase (Figure 9A). There was not much change in the G2/M phase but in the NS-398-treated cells there was subsequent cell accumulation in the G0/G1 phase, suggesting that COX-2 inhibitors prevent v-FLIP-HMVECs displaying anoikis from crossing the G1/S boundary. Similar results were obtained with celecoxib treatment but solvent treatment did not affect the cell cycle profile of v-FLIP-HMVECs displaying anoikis, further validating the specific effect of the drug used for treatment.
Effect of COX-2 inhibition on cell cycle of v-FLIP/K13-HMVECs during anoikis. (Aa–e) Propidium iodide (PI) staining of untreated, solvents for celecoxib or NS-398, celecoxib or NS-398-treated v-FLIP/K13-HMVEC-d cells was done. Data were analyzed using ModFit Lt V3 software (Verity Software House, Topsham, ME, USA). The percentages of cells at specific cell cycle phases are indicated and the numbers represent mean values of six independent experiments. The COX-2 inhibitors celecoxib and NS-398 induced a specific G0/G1 growth phase arrest as indicated by the accumulation of cells at this phase, with a corresponding decline in cells in the S (mitotic) phase. (f) The tabulated percentages are an average calculated from the results obtained from three independent experiments. (B) Schematic diagram depicting the multiple outcomes of v-FLIP/K13-induced COX-2 in ECs, the consequences and role in tumorigenesis. Results presented in this study show appreciable COX-2 gene expression, PGE2 secretion in v-FLIP/K13-HMVECs. Increased COX-2/PGE2 are involved in the upregulated secretion of inflammatory cytokines (MCP-1, RANTES, GRO-α/β, IL-8 and IL-6), inflammation-related genes, such as ICAM-1, VCAM-1 and chemokines, such as CXCL-6 and CXCL-5. v-FLIP/K13 induced the metastasis-related metalloprotease (MMP-10). In addition, COX-2 induction enhanced the phosphorylation of FAK, Src at the FAs in v-FLIP-HMVECs. FLIP/K13-induced COX-2/PGE2 also have important roles in the activation of attachment/ECM interaction-induced survival kinases (NF-κB, PI3 K and AKT), cell/cell contact-mediating protein laminin-γ and cytoskeletal rearrangement-linked small G-protein (Rac1-GTPase). Increased COX-2/PGE2 in v-FLIP-HMVECs conferred survival by providing resistance to anoikis. Cell survival cytokines, cell survival genes (cIAP-1, cIAP-2, xIAP, SOD2, BCL2A, IEX-1), antiapoptotic proteins (Bcl-2, MCL-1, BCL-xL, BIM), BAX translocation to the cytoplasm and anchorage-independent colony formation were upregulated. v-FLIP/K13-HMVECs enhanced the transitions of EndMT by reducing E-cadherin and upregulating the expression of slug, snail, twist and laminin-γ genes. In summary, KSHV oncogenic protein utilizes host cellular machinery and manipulates the cellular-inducible angiogenic stress response gene COX-2 and its lipid metabolite PGE2 to its advantage to enhance its transforming ability, metastatic potential and inflammatory phenotype, and to promote anoikis resistance and prolong cell survival.
Discussion
The biological effects of KSHV oncoprotein v-FLIP/K13, including antiapoptotis, NF-κB pathway activation, tumorigenesis, transformation and inflammation, have been well studied in KSHV-associated neoplasms.38, 39, 40 Similary, the potential of COX-2 pathway inhibitors in regulating the NF-κB pathway is also well established, but the connection between v-FLIP-induced oncogenesis and the COX-2 pathway has never been explored in detail. The central theme of our study was to evaluate the v-FLIP-COX-2-PGE2 connection, consequences and mechanism of COX-2 inhibition in v-FLIP-dependent growth, and potential of COX-2 inhibitors in v-FLIP-mediated pathogenesis. Since KS is an endothelial cell vascular neoplasm, we chose to work with endothelial (HMVEC-d) cells throughout this study. The novel findings (Figure 9B) of our study establish that (1) v-FLIP/K13 modulates transcriptional regulation of COX-2/m-PGES-1 and enhances PGE2 secretion. (2) KSHV latency protein LANA-1, which has been shown to be regulated at the transcriptional level by PGE2 and downregulated by COX-2 inhibition5, 7, 8, 9 did not upregulate COX-2/PGE2. (3) v-FLIP/K13 preferentially utilizes NF-κB to regulate COX-2 transcription. (4) COX-2 inhibition downregulated the v-FLIP/K13-mediated anoikis resistance. (5) COX-2 inhibition could also down-regulate the v-FLIP/K13 induced paracrine secreted factors required for maintaining anoikis resistance. (6) COX-2 inhibition reduced the potential of v-FLIP/K13-HMVECs for ECM interactions, mediated FAK/Src phosphorylation, reduced the number of the FAs and even reduced the size of FAs. (7) Attachment of COX-2 inhibitor-pretreated v-FLIP/K13-HMVEC cells could not induce NF-κB or AKT phosphorylation. (8) v-FLIP-mediated anoikis resistance in endothelial cells might be due to the enhanced levels of proteins such as MCL-1. (9) COX-2 inhibition accelerated anoikis in v-FLIP/K13-HMVECs, which could be due to the reduction in the number of cells in the ‘S’ phase of the cell cycle. We have addressed the mechanism of reduced anoikis resistance upon COX-2 inhibition in the v-FLIP/K13-HMVECs. Overall, our results allow new insights into the etiology of the molecular mechanisms granting anoikis resistance in KSHV oncoprotein-transduced ECs, inflammation and metastatic properties of v-FLIP/K13. This opens new avenues to pharmacological intervention of KS tumors with FDA-approved COX-2 inhibitors.
NFκB activation seems to be the pivotal link between v-FLIP and COX-2/PGE2 induction. Bay 11-7082, a known pharmacological inhibitor of NFκB activation, could inhibit v-FLIP-mediated COX-2 induction, whereas NFAT and CREB inhibition could not. Another interesting observation of our study is the activation of various MAPKs (p38γ, p38γ, AKT-Pan, AKT1, RSK1 and GSK3) upon v-FLIP introduction into endothelial cells. Inhibitors of AKT, p38 and RSK1 downregulated COX-2 gene expression, COX-2 protein levels and PGE2 secretion, suggesting that v-FLIP utilizes signaling pathways to maintain stress response angiogenic protein COX-2.
K13/v-FLIP has been previously reported to induce the expression of various proinflammatory cytokines and growth factors. We additionally found that factors having a pivotal role in inflammation, neo-vascularization, chemo-attraction, host defense, development and homeostasis of the immune system such as IL-8/CXCL8, IL-6, CXCLs 5 and 6, CCLs 2 and 20 can be effectively downregulated upon COX-2 inhibitor treatment. Higher levels of adhesion molecules such as ICAM-1 and VCAM-1 were expressed in v-FLIP/K13-HMVECs, which might synergize with chemokines and their ligands to enhance the recruitment of inflammatory and blood cells into KS lesions, thus the arrest of infected, transformed cells would help in survival and growth. We observed significantly decreased VCAM-1 gene expression in the cells pretreated with COX-2 inhibitors that might be due to the downregulation of NF-κB leading to reduced binding of NF-κB on the promoter region of VCAM-1. Celecoxib or NS-398 treatment could also abrogate p65 nuclear translocation and p65 DNA-binding activity in the v-FLIP/K13-HMVEC cells. As NF-κB pathway-dependent genes are involved in the transformation, cancer cell migration, tissue invasion and metastasis, and antiapoptosis and anchorage-independent growth, we assessed the role of COX-2 inhibitors in regulating these processes. Consistent with downregulation of inflammatory cytokines and adhesion molecule genes, celecoxib or NS-398 could abrogate the levels of total and active MMP-10 required for cell invasion. Though we show that celecoxib or NS-398 treatment could abrogate p65 nuclear translocation and p65 DNA-binding activity in v-FLIP/K13-HMVEC cells (Figure 4), the mechanism by which COX-2 inhibitors block NF-κB has not been deciphered and will be a question for our future studies. At present, we can predict multiple scenarios controlling COX2 inhibitor-mediated NF-κB block, such as PGE2 secretion per se or the cytokines and spindle cell differentiation induced by v-FLIP transduction. (1) Cells treated with COX-2 inhibitors exhibit reduced COX-2 and secrete lower levels of PGE2. Although PGE2 has short half-life, it can mediate its downstream signaling events through binding to eicosanoid receptors, and this binding has been shown to initiate an avalanche of temporal effects on cellular signaling such as Src, PI3K, PKCζ/γ, NF-κB and Ca2+.7 PGE2 has been shown to be an important factor involved in establishing and sustaining inflammation, and much of its activity has been shown to be dependent on its ability to induce NF-κB in other systems as suggested in our previous study.7 (2) Exogenous PGE2 has also been shown to enhance the activity of multiple transcription factors,7 which might be involved in maintaining the NF-κB promoter activation/transcriptional regulation. (3) Role of secreted cytokines upon v-FLIP transduction in maintaining the levels of NF-κB cannot be ruled out. Reduced secretion of cytokines along with lowered spindle cell differentiation upon COX-2 inhibitor pretreatment could affect the activity status of NF-κB. These secreted factors might be potentially involved in the activation of NF-κB.
This is the first report suggesting v-FLIP's key role in cytoskeletal rearrangement modulating kinases such as AKT, src, laminin-γ-1 and Rac1-GTPases. How cytoskeletal alterations are engendered remains unknown, but it is clear that COX-2/PGE2/NF-κB activation is responsible for activating major survival kinases, thereby contributing to v-FLIP's known antiapoptotic effects. v-FLIP-expressing endothelial cells expressed a higher level of integrin-linked kinase, an ankyrin repeat containing serine–threonine protein kinase interacting with the cytoplasmic domains of β-integrins and numerous cytoskeleton-associated proteins, which probably functions in cell adhesion, cell shape changes and deposition of ECM as shown in a number of human malignancies.41
KSHV infection or v-FLIP or LANA-1 expression in primary HMVEC-d cells has been shown42, 43, 44 to induce a spindle cell conversion phenotype. The infected ECs develop a spindle shape resembling that of KS lesion cells and show characteristics of a transformed phenotype including loss of contact inhibition and acquisition of anchorage-independent growth.44 Nodular KS lesions contain latently infected spindle cells and demonstrate monoclonal, oligoclonal and polyclonal patterns of infection, implying that KSHV infection precedes tumor expansion. Inhibition of anoikis is a prerequisite for infection progression and metastasis. v-FLIP in these latently infected cells in nodular lesions has been proposed to inhibit anoikis which will allow the detachment and spreading of single cells, and contributes to the metastasis recognized as a hallmark of epidemic KS resulting in the development of larger monoclonal lesions.45 Therefore, we focused our study on understanding the involvement of COX-2/PGE2 in v-FLIP-induced anoikis resistance. v-FLIP rescued HMVEC-d cells from anoikis and it was predominantly dependent on the level of COX-2/PGE2. v-FLIP-mediated anoikis resistance was partly attributable to secreted survival factors, which were found to be reduced upon COX-2 inhibitor treatment. Though COX-2/PGE2 has previously been reported to activate prosurvival pathways including Raf-MEK-ERK1/2, PI3K/AKT, cAMP/protein kinase A and Wnt/β-catenin, it is still unclear how PGE2 signaling couples to the intrinsic cell death machinery. Clinically relevant downstream mediators of COX-2/PGE2-mediated apoptosis suppression and COX-2 inhibitor-induced apoptosis remain elusive. We expanded our study to understand the probable mechanism of v-FLIP-induced COX-2/PGE2-dependent anoikis resistance along with the potential of COX-2 inhibitors in inducing anoikis. As anoikis execution can be controlled by BH3-only proteins (Bid, Bad, Bim, Bik, Bmf and Noxa) and Bim translocation to mitochondria and its rapid interaction with Bcl-XL and neutralization of prosurvival function and promoting Bax activation, we evaluated these events in untreated and COX-2 inhibitor-treated endothelial cells. COX-2 inhibitor treatment not only inhibited the expression of cellular prosurvival proteins including cIAP-1, cIAP-2, x-IAP and BCL2A1 but also reduced the level of antioxidant SOD2 expression.
The life–death switch of the intrinsic pathway of apoptosis is governed in multiple ways such as: (1) damage-sensing BH3-only members of the Bcl-2 family, which reside upstream of proapoptotic proteins Bax and Bak46 and promote apoptosis by antagonizing the antiapoptotic activity of prosurvival Bcl-2 family members; (2) release of cytochrome c from the mitochondria, (3) altered localization of the proapoptotic protein BAX, (4) direct activation of Bax or Bak47 and the balance between proapoptotic (Bax and Bak) and antiapoptotic (Bcl-xL and Mcl-1) portions of the Bcl-2 proteins.48 Direct activation of Bax has been shown by Bim, the most powerful BH3-only protein with tumor-suppressor activity. Bim has also been reported as a key mediator of apoptosis and a quick responder to several cancer therapeutics including targeted therapies in ‘oncogene-addicted’ cancer cell lines.49 Celecoxib and NS-398 treatment induced the levels of Bim that might be due to the downregulation of COX-2 and the subsequently diminished pool of PGE2. PGE2 signaling has been shown to promote ERK1/2-dependent Bim phosphorylation and proteasomal degradation, thus leading to Bim repression and apoptosis suppression in human colorectal adenoma cells. Integrin expression and enhanced ECM interaction of v-FLIP-transduced cells might be promoting kinases such as MAPKs and PI3K/AKT, which can mediate phosphorylation of Bim, culminating in its proteasome-dependent degradation and thus prolonged cell survival. Upon COX-2 inhibition, downregulated MAPKs and PI3K/AKT signaling might be converging to reduce Bim phosphorylation, leading to the abolishment of its degradation and finally to its accumulation, thus promoting faster cell death. Another possibility is the involvement of v-FLIP-induced ROS, which can induce direct oxidation of the tyrosine kinase Src, thereby leading to its activation. This activated kinase in turn might be eliciting activation of MAPKs and PI3K/AKT signaling pathways leading to Bim phosphorylation/degradation, thus contributing to prolonged cell survival.
Previous studies have found massive inflammation and dysregulated angiogenesis in KS lesions accompanied by a mesenchymal phenotype in KS tumor cells.50 These studies suggested a role of EndMT during KS lesion development; therefore, we focused on the role of v-FLIP/K13-induced COX-2/PGE2 in EndMT signatures (slug, snail, twist, laminin-γ and E-cadherin). Additionally, COX-2 has been demonstrated to regulate snail and E-cadherin51 expression in non-small-cell lung carcinoma and colon cancer cells. Twist has been shown to act independently of snail to repress E-cadherin and to upregulate fibronectin and N-cadherin.52 COX-2 inhibition effectively reduced the expression of slug, snail, twist and laminin-γ but enhanced the expression of E-cadherin in v-FLIP-transformed endothelial cell colonies. COX-2 inhibition might be acting at the level of controlling the levels of TGF-β1 secretion. TGF-β1 has been shown to induce EndMT and KSHV infection, and v-FLIP has been shown to upregulate TGF-β1 that could be effectively downregulated by COX-2 inhibition in the endothelial cells.6
Studies to understand the role of v-FLIP-induced COX-2/PGE2 in DNA–protein interactions, activation of various transcription factors, nuclear signaling of various survival kinases, PGE2 receptors (eicosanoid receptors) and their expression, changes in plasticity biomarkers of EndMT useful for the recognition of EC-spindle cell phenotypic transitions using a panel of human lymphoma cell lines harboring KSHV and expressing v-FLIP are ongoing in our lab. Collectively, our studies indicate the promising clinical perspective of COX-2 inhibitors (celecoxib and NS-398) and show that COX-2/PGE2 promote v-FLIP-expressing endothelial cell survival, cell invasion, cellular transformation and resistance to cell death upon detachment from the ECM. Our results showing the induction of DR5 expression upon celecoxib treatment are very promising and are currently being studied further as this appears to be potentially significant chemotherapeutically.
Materials and methods
Cell culture
Human microvascular dermal ECs of passages 5–7 (HMVEC-d, CC-2543; Lonza Walkersville, Walkersville, MD, USA) were grown in endothelial basal media-2 with growth factors (Lonza Walkersville). All stock preparations of purified KSHV were monitored for endotoxin contamination by standard Limulus assay (Limulus amebocyte lysate endochrome; Charles River Endosafe, Charleston, SC, USA).5 All cells were cultured in lipopolysaccharide-free medium.
Reagents
COX-2-specific inhibitors NS-398 (N-(2-cyclohexyloxy-4-nitrophenyl)-methanesulfonamide) and celecoxib (4-(5-(4-Methylphenyl)-3-(trifluoromethyl)-1H-pyrazol-1-yl)-benzene sulfonamide) were purchased from Calbiochem (La Jolla, CA, USA) and Tocris Biosciences (Ellisville, MO, USA), respectively. The COX-2 luciferase reporter plasmids containing the 5′ flanking region of human COX-1 and COX-2 promoter deletion and mutation constructs linked to luciferase reporter plasmids were obtained from Dr M Fresno (Madrid, Spain) and have been described previously.17 VIVIT, a plasmid containing the NFAT regulatory domain, was obtained from Addgene (Cambridge, MA, USA) and has been described previously.8
Real-time RT–PCR
Transcripts of the genes of interest were detected by real-time RT–PCR using gene-specific primers (Table 1) as per procedures described previously.5
ELISA for PGE2 detection
Levels of PGE2 in the supernatants of lentivirus-transduced ECs were measured by a commercially available enzyme immunoassay as described previously.5, 6
Transient transfections
For reporter gene assays, 293 cells were transfected using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) as described before.8
Cytotoxicity assay
Supernatants were collected from the target cells incubated with DMEM containing different concentrations of various inhibitors for the indicated time points, and cellular toxicity was estimated using a cytotoxicity assay kit (Promega, Madison, WI, USA).5
Luciferase reporter assays
The effect of v-FLIP/K13 on the COX-2 full-length promoter (P2-1900), its deletion constructs and constructs with mutant transcription factor binding sites17 and the COX-1 promoter (P2-1009) was measured as described previously.8
Lentivirus production, infection of HMVEC-d cells and testing lentiviral transduction
Lentiviral infection was done as described by Vart et al.53 Vesicular stomatitis virus-G envelope-pseudotyped lentiviruses expressing viral genes (v-FLIP/K13/ORF71, v-cyclin/ORF72, LANA-1/ORF73 and K12) were produced using a four-plasmid transfection system.6 Lentiviruses expressing pSIN (empty vector) or GFP were used as control. Total RNA was extracted from lentiviral-transduced HMVECs and subjected to DNase I digestion (Invitrogen). This RNA was subsequently used for cDNA synthesis and real-time qRT–PCR was done for viral (ORF73, ORF50) and host genes (COX-1, COX-2 and m-PGES-1). The cells were split at 24 h after lentivirus infection and allowed to grow until 80–90% confluent. These cells were serum starved, and supernatants were collected after 48 h, spun at 1000 r.p.m. for 10 min at 4°C to remove particulates, and used for measuring PGE2 levels by ELISA.5
Preparation of nuclear extracts
p-SIN, GFP or v-FLIP/K13-HMVEC-d cells were harvested to prepare nuclear extracts as described,8 purity of the nuclear extracts was assessed by immunoblotting using anti-lamin B antibodies, and cytoskeletal contamination was checked using an anti-β tubulin antibody.
RhoA-GTPase and Rac1-GTPase activity by G-LISA activation assay
The degree of RhoA-GTPase and Rac-1-GTPase activation in various lysates was determined by RhoA and Rac-1 G-LISA from Cytoskeleton, Inc. as described in our previous publication.6
Signaling inhibitor treatment
For luciferase assays, gene expression, PGE2 secretion and COX-2/COX-1/Tubulin protein measurements, cells were pretreated with either solvent controls or signaling inhibitors. Briefly, target cells grown to confluence in 25-cm2 flasks were serum starved for 10 h, exposed to either solvent controls or signaling inhibitors such as U0126, SB203580, SB216763, AKT inhibitor API-2 (Triciribine; selective inhibitor of Akt signaling), SL0101-1 (3-((3,4-Di-O-acetyl-6-deoxy-a-L-mannopyranosyl) oxy)-5, 7-dihydro-2-(4-hydroxyphenyl)-4H-1benzopyran-4-one; selective inhibitor of p90 ribosomal S6 kinase (RSK), or 5 μM Bay 11-7082 for 1 h at 37°C before transfection with v-FLIP/K13 expression plasmid.
Transcription factor activation assay
A total of 2 μg of each nuclear extract was used for measuring NFAT, CREB and phospho-CREB activity using ELISA-based TransAM-NFAT and TransAM-CREB kits (Active Motif Corp., Carlsbad, CA, USA) as described previously.8
Electrophoretic mobility shift assay
Consensus double-strand oligo-deoxynucleotide probes for NF-κB (5′-TAGTTGAGGGGACTTTCCCAGGC-3′) were radioactively labeled using [γ-32P] ATP and T4 polynucleotide kinase as described previously.8 Then, 2 μg of nuclear extract was incubated with a γ-32P-labeled double-stranded oligonucleotide probe, the binding reaction was carried out, bound complexes were separated by polyacrylamide gel electrophoresis and visualized by autoradiography as described previously.8 A competition EMSA was performed by adding a 100-fold molar excess of unlabeled double-stranded oligonucleotide cold probes.
Phospho-MAPK antibody arrays
p-SIN or v-FLIP/K13-HMVEC lysates were used for analysis of the phosphorylation state of all MAPKs using a human phospho-MAPK array kit (R&D Systems, Minneapolis, MN, USA) as described before.8
Measurement of GSK3β activity by FACE GSK3β ELISA
Phospho and total-GSK3β levels were measured in p-SIN or v-FLIP/K13-HMVECs by FACE (fast activated cell-based) ELISA kit (Active Motif Corp.) as described previously.8
Western blot analysis
Cell extracts were quantitated by BCA protein assay, and then equal amounts of protein (20 μg/lane) were separated by SDS-PAGE and electrotransferred to 0.45-μm nitrocellulose membranes. The membranes were blocked with 5% BSA, probed with specific antibodies and visualized using an ECL detection system.5
Cell number and viability assays
The in vitro effects of COX-2 inhibition on cell numbers and viability were determined by traditional Trypan blue staining (evaluation of cell membrane integrity) and the quantitation of the number of viable cells with their metabolically active mitochondria (an index of cell proliferation) by the MTT-based colorimetric assay (ATCC, Manassas, VA, USA) as described previously.6
Anoikis induction
The anoikis assay was done using the CytoSelect 96-well anoikis assay kit from Cell Biolabs, Inc. (San Diego, CA, USA). Briefly, cells were untreated or treated with COX-2 inhibitors or their respective solvent controls, then 100 μl of each cell suspension (2.0 × 106 cells/ml) in the presence of culture medium or supernatant from p-SIN or v-FLIP/K13-HMVECs were added to the anchorage-resistant plate or a control 96-well cell culture plate. Cells were cultured for 12 h at 37°C and 5% CO2. Analysis of cell viability was done in multiple ways, such as MTT (colorimetric) and Calcein AM (485 nm/515 nm) fluorimetric detection. Calcein AM was used as a fluorescent marker of cell viability as it labels viable cells. Anoikis propelled cell death was measured by Ethidium homodimer (EthD-1), an excellent marker for measuring dead cells. EthD-1 is a red fluorescent dye that can only penetrate damaged cell membranes. EthD-1 fluoresces with a 40-fold enhancement upon binding ssDNA, dsDNA, RNA, oligonucleotides and triplex DNA. Background fluorescence levels are very low because the dyes are virtually non-fluorescent before interacting with cells. Cells were also monitored microscopically for the presence of the green Calcein AM (Ex: 485 nm and Em: 515 nm) or red EthD-1 (Ex: 525 nm and Em: 590 nm) fluorescence.
MMPs activity assay
Levels of total and active MMPs (−2, −9 and −10) in the conditioned media obtained from p-SIN or v-FLIP/K13-HMVECs cells untreated or pretreated with NS-398 or celecoxib for 4 h, 8 h, and 24 h were detected by ELISA as described in our previous publication.6
Invasion assays
Effect of COX-2 inhibitor treatment on the intrinsic invasive potential of v-FLIP/K13-HMVECs and paracrine secretion mediated invasion through the ECM was quantitated by the Innocyte cell invasion assay as described previously.6
Cell transformation assay
v-FLIP-HMVEC cells were used to evaluate the effect of celecoxib or NS-398 or their solvent controls on the colony-forming capacity of v-FLIP-HMVECs and to measure their proliferative capacity within the colonies as per the manufacturer's guidelines using an MTT-based CytoSelect cell transformation assay (Cell Biolabs, Inc., San Diego, CA, USA).
Cell cycle analysis by flow cytometry
v-FLIP/K13-HMVECs were either untreated or treated with drugs (NS-398 or celecoxib) or solvent control as described for the cell number and viability assays. Harvested cells were diluted to contain ∼106 cells/ml, and DNA distribution analysis was performed as described in our previous publication.6
Caspase activity assay
Caspase (3, 8 and 9) activities were measured in cell lysates by caspase-Glo-3, caspase-Glo-8 and caspase-Glo-9 assays from Promega as per the manufacturer's instructions.
Mn-SOD activity assay
Mn-SOD activity was measured using a superoxide dismutase assay kit (Cayman's chemical, Ann Arbor, MI, USA) as per manufacturer's instructions. This kit utilizes a tetrazolium salt for the detection of superoxide radicals generated by xanthine oxidase and hypoxanthine. One unit of SOD is defined as the amount of enzyme needed to exhibit 50% dismutation of the superoxide radical. This kit provides a means to accurately quantify the activity of all three types of SOD (Cu/Zn, Mn and Fe-SOD). Cells harvested using rubber policeman were pelleted at 1000 g for 10 min at 4°C. Cell pellets were sonicated in ice cold 20 mM HEPES, pH 7.2 containing 1 mM EGTA, 210 mM mannitol, 70 mM sucrose, centrifuged at 1500 × g for 5 min at 4°C. Supernatants were used for assaying total SOD activity (cytosolic and mitochondrial). To separate the two enzymes, the 1500 × g supernatant was centrifuged at 10 000 × g for 15 min at 4°C. The resulting supernatant was used to assay cytosolic SOD whereas the pellet was used for mitochondrial SOD. The mitochondrial pellet was homogenized in ice cold 20 mM HEPES, pH 7.2 containing 1 mM EGTA, 210 mM mannitol, 70 mM sucrose and the supernatant was used for assay. Potassium cyanide (1–3 mM) was added to the assay to inhibit both Cu/Zn-SOD and extracellular SOD, resulting in the detection of MnSOD only.
Transcription factor activation assay
A total of 5 μg of each nuclear extract was used for NF-κB activation using an enzyme-linked immunosorbent assay (ELISA)-based TransAM NF-κB Family kit (Active Motif Corp.) per the manufacturer's instructions. A competition assay was done by premixing nuclear extracts for 30 min at 4 °C with wild-type consensus and mutated consensus oligonucleotides provided in the kit before addition to the probe immobilized on the plate.
Abbreviations
- COX-2:
-
cyclooxygenase-2
- ECM:
-
extracellular matrix
- HMVEC:
-
human microvascular endothelial cells
- ICAM-1:
-
intercellular adhesion molecule-1
- PGE2:
-
prostaglandin E2
- VCAM-1:
-
vascular cell adhesion molecule-1
- v-FLIP/K13:
-
FADD-like interferon converting enzyme or caspase 8 (FLICE) inhibitory protein.
References
Cesarman E, Chang Y, Moore PS, Said JW, Knowles DM . Kaposi's sarcoma-associated herpesvirus-like DNA sequences in AIDS-related body-cavity-based lymphomas. N Engl J Med 1995; 332: 1186–1191.
Cesarman E, Knowles DM . The role of Kaposi's sarcoma-associated herpesvirus (KSHV/HHV-8) in lymphoproliferative diseases. Semin Cancer Biol 1999; 9: 165–174.
Boshoff C, Schulz TF, Kennedy MM, Graham AK, Fisher C, Thomas A et al. Kaposi's sarcoma-associated herpesvirus infects endothelial and spindle cells. Nat Med 1995; 1: 1274–1278.
Boshoff C, Weiss R . AIDS-related malignancies. Nat Rev Cancer 2002; 2: 373–382.
Sharma-Walia N, Raghu H, Sadagopan S, Sivakumar R, Veettil MV, Naranatt PP et al. Cyclooxygenase 2 induced by Kaposi's sarcoma-associated herpesvirus early during in vitro infection of target cells plays a role in the maintenance of latent viral gene expression. J Virol 2006; 80: 6534–6552.
Sharma-Walia N, Paul AG, Bottero V, Sadagopan S, Veettil MV, Kerur N et al. Kaposi's sarcoma associated herpes virus (KSHV) induced COX-2: a key factor in latency, inflammation, angiogenesis, cell survival and invasion. PLoS Pathog 2010; 6: e1000777.
George Paul A, Sharma-Walia N, Kerur N, White C, Chandran B . Piracy of prostaglandin E2/EP receptor-mediated signaling by Kaposi's sarcoma-associated herpes virus (HHV-8) for latency gene expression: strategy of a successful pathogen. Cancer Res 2010; 70: 3697–3708.
Sharma-Walia N, Paul AG, Patel K, Chandran K, Ahmad W, Chandran B . NFAT and CREB regulate Kaposi's sarcoma associated herpes virus (KSHV) induced cyclooxygenase-2 (COX-2). J Virol 2010; 84: 12733–12753.
Paul AG, Sharma-Walia N, Chandran B . Targeting KSHV/HHV-8 latency with COX-2 selective inhibitor nimesulide: a potential chemotherapeutic modality for primary effusion lymphoma. PLoS One 2011; 6: e24379.
Sun Q, Matta H, Lu G, Chaudhary PM . Induction of IL-8 expression by human herpesvirus 8 encoded vFLIP K13 via NF-kappaB activation. Oncogene 2006; 25: 2717–2726.
Grossmann C, Podgrabinska S, Skobe M, Ganem D . Activation of NF-kappaB by the latent vFLIP gene of Kaposi's sarcoma-associated herpesvirus is required for the spindle shape of virus-infected endothelial cells and contributes to their proinflammatory phenotype. J Virol 2006; 80: 7179–7185.
Punj V, Matta H, Schamus S, Chaudhary PM . Integrated microarray and multiplex cytokine analyses of Kaposi's sarcoma associated herpesvirus viral FLICE inhibitory protein K13 affected genes and cytokines in human blood vascular endothelial cells. BMC Med Genomics 2009; 2: 50.
Sakakibara S, Pise-Masison CA, Brady JN, Tosato G . Gene regulation and functional alterations induced by Kaposi's sarcoma-associated herpesvirus-encoded ORFK13/vFLIP in endothelial cells. J Virol 2009; 83: 2140–2153.
Rennebeck G, Martelli M, Kyprianou N . Anoikis and survival connections in the tumor microenvironment: is there a role in prostate cancer metastasis? Cancer Res 2005; 65: 11230–11235.
Efklidou S, Bailey R, Field N, Noursadeghi M, Collins MK . vFLIP from KSHV inhibits anoikis of primary endothelial cells. J Cell Sci 2008; 121 (Part 4): 450–457.
Casanova I, Parreno M, Farre L, Guerrero S, Cespedes MV, Pavon MA et al. Celecoxib induces anoikis in human colon carcinoma cells associated with the deregulation of focal adhesions and nuclear translocation of p130Cas. Int J Cancer 2006; 118: 2381–2389.
Iniguez MA, Martinez-Martinez S, Punzon C, Redondo JM, Fresno M . An essential role of the nuclear factor of activated T cells in the regulation of the expression of the cyclooxygenase-2 gene in human T lymphocytes. J Biol Chem 2000; 275: 23627–23635.
Jiang XH, Lam SK, Lin MC, Jiang SH, Kung HF, Slosberg ED et al. Novel target for induction of apoptosis by cyclo-oxygenase-2 inhibitor SC-236 through a protein kinase C-beta(1)-dependent pathway. Oncogene 2002; 21: 6113–6122.
He Q, Luo X, Jin W, Huang Y, Reddy MV, Reddy EP et al. Celecoxib and a novel COX-2 inhibitor ON09310 upregulate death receptor 5 expression via GADD153/CHOP. Oncogene 2008; 27: 2656–2660.
Penning TD, Talley JJ, Bertenshaw SR, Carter JS, Collins PW, Docter S et al. Synthesis and biological evaluation of the 1,5-diarylpyrazole class of cyclooxygenase-2 inhibitors: identification of 4-[5-(4-methylphenyl)-3-(trifluoromethyl)-1H-pyrazol-1-yl]benze nesulfonamide (SC-58635, celecoxib). J Med Chem 1997; 40: 1347–1365.
Half EE, Arber N . Chemoprevention of colorectal cancer: two steps forward, one step back? Future Oncol 2006; 2: 697–704.
Reddy BS, Hirose Y, Lubet R, Steele V, Kelloff G, Paulson S et al. Chemoprevention of colon cancer by specific cyclooxygenase-2 inhibitor, celecoxib, administered during different stages of carcinogenesis. Cancer Res 2000; 60: 293–297.
Meredith JE Jr, Fazeli B, Schwartz MA . The extracellular matrix as a cell survival factor. Mol Biol Cell 1993; 4: 953–961.
Frisch SM, Ruoslahti E . Integrins and anoikis. Curr Opin Cell Biol 1997; 9: 701–706.
Obara S, Nakata M, Takeshima H, Katagiri H, Asano T, Oka Y et al. Integrin-linked kinase (ILK) regulation of the cell viability in PTEN mutant glioblastoma and in vitro inhibition by the specific COX-2 inhibitor NS-398. Cancer Lett 2004; 208: 115–122.
Ghosh S, Karin M . Missing pieces in the NF-kappaB puzzle. Cell 2002; 109 (Suppl): S81–S96.
Dogic D, Eckes B, Aumailley M . Extracellular matrix, integrins and focal adhesions. Curr Top Pathol 1999; 93: 75–85.
Katz BZ, Zamir E, Bershadsky A, Kam Z, Yamada KM, Geiger B . Physical state of the extracellular matrix regulates the structure and molecular composition of cell-matrix adhesions. Mol Biol Cell 2000; 11: 1047–1060.
Hynes RO . Integrins: versatility, modulation, and signaling in cell adhesion. Cell 1992; 69: 11–25.
Burridge K, Wennerberg K . Rho and Rac take center stage. Cell 2004; 116: 167–179.
Sharma-Walia N, Naranatt PP, Krishnan HH, Zeng L, Chandran B . Kaposi's sarcoma-associated herpesvirus/human herpesvirus 8 envelope glycoprotein gB induces the integrin-dependent focal adhesion kinase-Src-phosphatidylinositol 3-kinase-rho GTPase signal pathways and cytoskeletal rearrangements. J Virol 2004; 78: 4207–4223.
Nishida K, Kaziro Y, Satoh T . Anti-apoptotic function of Rac in hematopoietic cells. Oncogene 1999; 18: 407–415.
Arciniegas E, Neves CY, Carrillo LM, Zambrano EA, Ramirez R . Endothelial-mesenchymal transition occurs during embryonic pulmonary artery development. Endothelium 2005; 12: 193–200.
Leong KG, Niessen K, Kulic I, Raouf A, Eaves C, Pollet I et al. Jagged1-mediated Notch activation induces epithelial-to-mesenchymal transition through Slug-induced repression of E-cadherin. J Exp Med 2007; 204: 2935–2948.
Romano LA, Runyan RB . Slug is a mediator of epithelial-mesenchymal cell transformation in the developing chicken heart. Dev Biol 1999; 212: 243–254.
Bai J, Cederbaum AI . Catalase protects HepG2 cells from apoptosis induced by DNA-damaging agents by accelerating the degradation of p53. J Biol Chem 2003; 278: 4660–4667.
Liu X, Yue P, Zhou Z, Khuri FR, Sun SY . Death receptor regulation and celecoxib-induced apoptosis in human lung cancer cells. J Natl Cancer Inst 2004; 96: 1769–1780.
Chaudhary PM, Jasmin A, Eby MT, Hood L . Modulation of the NF-kappa B pathway by virally encoded death effector domains-containing proteins. Oncogene 1999; 18: 5738–5746.
Liu L, Eby MT, Rathore N, Sinha SK, Kumar A, Chaudhary PM . The human herpes virus 8-encoded viral FLICE inhibitory protein physically associates with and persistently activates the Ikappa B kinase complex. J Biol Chem 2002; 277: 13745–13751.
Matta H, Chaudhary PM . Activation of alternative NF-kappa B pathway by human herpes virus 8-encoded Fas-associated death domain-like IL-1 beta-converting enzyme inhibitory protein (vFLIP). Proc Natl Acad Sci USA 2004; 101: 9399–9404.
Hannigan G, Troussard AA, Dedhar S . Integrin-linked kinase: a cancer therapeutic target unique among its ILK. Nat Rev Cancer 2005; 5: 51–63.
Carroll PA, Brazeau E, Lagunoff M . Kaposi's sarcoma-associated herpesvirus infection of blood endothelial cells induces lymphatic differentiation. Virology 2004; 328: 7–18.
Ciufo DM, Cannon JS, Poole LJ, Wu FY, Murray P, Ambinder RF et al. Spindle cell conversion by Kaposi's sarcoma-associated herpesvirus: formation of colonies and plaques with mixed lytic and latent gene expression in infected primary dermal microvascular endothelial cell cultures. J Virol 2001; 75: 5614–5626.
Moses AV, Fish KN, Ruhl R, Smith PP, Strussenberg JG, Zhu L et al. Long-term infection and transformation of dermal microvascular endothelial cells by human herpesvirus 8. J Virol 1999; 73: 6892–6902.
Osmond DH, Buchbinder S, Cheng A, Graves A, Vittinghoff E, Cossen CK et al. Prevalence of Kaposi sarcoma-associated herpesvirus infection in homosexual men at beginning of and during the HIV epidemic. JAMA 2002; 287: 221–225.
Cheng EH, Wei MC, Weiler S, Flavell RA, Mak TW, Lindsten T et al. BCL-2, BCL-X(L) sequester BH3 domain-only molecules preventing BAX- and BAK-mediated mitochondrial apoptosis. Mol Cell 2001; 8: 705–711.
Gallenne T, Gautier F, Oliver L, Hervouet E, Noel B, Hickman JA et al. Bax activation by the BH3-only protein Puma promotes cell dependence on antiapoptotic Bcl-2 family members. J Cell Biol 2009; 185: 279–290.
Youle RJ, Strasser A . The BCL-2 protein family: opposing activities that mediate cell death. Nat Rev Mol Cell Biol 2008; 9: 47–59.
Balmanno K, Cook SJ . Tumour cell survival signalling by the ERK1/2 pathway. Cell Death Differ 2009; 16: 368–377.
Greene W, Kuhne K, Ye F, Chen J, Zhou F, Lei X et al. Molecular biology of KSHV in relation to AIDS-associated oncogenesis. Cancer Treat Res 2007; 133: 69–127.
Dohadwala M, Yang SC, Luo J, Sharma S, Batra RK, Huang M et al. Cyclooxygenase-2-dependent regulation of E-cadherin: prostaglandin E(2) induces transcriptional repressors ZEB1 and snail in non-small cell lung cancer. Cancer Res 2006; 66: 5338–5345.
Yang Z, Zhang X, Gang H, Li X, Li Z, Wang T et al. Up-regulation of gastric cancer cell invasion by Twist is accompanied by N-cadherin and fibronectin expression. Biochem Biophys Res Commun 2007; 358: 925–930.
Vart RJ, Nikitenko LL, Lagos D, Trotter MW, Cannon M, Bourboulia D et al. Kaposi's sarcoma-associated herpesvirus-encoded interleukin-6 and G-protein-coupled receptor regulate angiopoietin-2 expression in lymphatic endothelial cells. Cancer Res 2007; 67: 4042–4051.
Acknowledgements
This study was supported in part by the Public Health Service Grant CA128560 to NSW, the Rosalind Franklin University of Medicine and Science start up fund and the H. M. Bligh Cancer Research Fund. The funders had no role in study design, data collection, analysis and decision to publish, or preparation of the manuscript. We gratefully acknowledge Dr Alice Gilman-Sach's suggestions in writing this manuscript and thank Keith Philibert for critically reading this manuscript. We gratefully acknowledge Drs. Iñiguez MA, Preet Chaudhary, Mary Collins and Axel H. Schönthal for providing COX-2 and COX-1 promoter constructs, v-FLIP-transduced cells, v-FLIP/K13 antibody and dimethyl celecoxib, respectively. We highly appreciate help from Waseem Ahmad (summer student) in standardizing dilutions for several antibodies used in the study. We also thank Bob Dickinson for his technical expertise in using the FACS facility in the Microbiology and Immunology department at RFUMS.
Author contributions: NSW conceived, designed and performed most of the experiments. NSW analyzed results, performed statistical analyses, compiled all the results, wrote the manuscript, and is the corresponding author. KP performed WB. KC performed cell cycle analysis and staining of death receptors by FACS. AM performed WB for all the experiments done for manuscript revision whereas VB prepared v-FLIP lentivirus and contributed to promoter work. NK and AP performed promoter analysis.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on the Oncogenesis website
Rights and permissions
This work is licensed under the Creative Commons Attribution-NonCommercial-No Derivative Works 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/3.0/
About this article
Cite this article
Sharma-Walia, N., Patel, K., Chandran, K. et al. COX-2/PGE2: molecular ambassadors of Kaposi's sarcoma-associated herpes virus oncoprotein-v-FLIP. Oncogenesis 1, e5 (2012). https://doi.org/10.1038/oncsis.2012.5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/oncsis.2012.5
Keywords
This article is cited by
-
Identification of upstream miRNAs of SNAI2 and their influence on the metastasis of gastrointestinal stromal tumors
Cancer Cell International (2019)
-
Herpesvirus-encoded GPCRs: neglected players in inflammatory and proliferative diseases?
Nature Reviews Drug Discovery (2014)