Abstract
Claudins are integral tight junction proteins that are responsible for maintaining the integrity of epithelial cell architecture and cell polarity. Claudin-3 and -4 are overexpressed in several cancers and have been shown to act as receptors for the Clostridium perfringens enterotoxin (CPE), a toxin that causes rapid cell lysis. CPE has demonstrated effectiveness in treating several different cancers in mouse models, provided that these cancers express claudin-3 or claudin-4. Here, we show that claudin-3/4 expression is not an absolute requirement for CPE action and, through overexpression and knockdown experiments, we identify claudin-6 as a novel functional receptor for CPE. Indeed, UCI-101, an ovarian cancer cell line highly sensitive to CPE, does not express claudin-3/4 and knockdown of claudin-6 in these cells decreases CPE sensitivity. Moreover, two different ovarian cell lines that are resistant to the effects of CPE can be made sensitive through claudin-6 overexpression. Binding assays show that CPE can indeed bind claudin-6 in cells and that this binding is associated with CPE cytotoxicity. Multicellular tumor spheroids experiments demonstrate that claudin-6 can also be a target of CPE in three-dimensional cultures. Our data establish claudin-6 as a novel receptor for CPE and introduces the possibility of a novel targeted therapeutic for ovarian and other cancers that express claudin-6.
Similar content being viewed by others
Introduction
Ovarian cancer is the seventh most common cancer in women worldwide1 and remains one of the most difficult cancers to diagnose early and treat effectively.2 Standard treatment for ovarian cancer consists of cytoreductive surgery followed by a combination of platinum and taxane-based therapy.3 Unfortunately, many patients present with tumors intrinsically resistant to this regimen, while the sensitive tumors often develop drug resistance,3 resulting in a 5-year survival below 35%. Over the last few years, there has been a significant increase in our understanding of the underlying molecular mechanisms leading to epithelial ovarian cancer, but in spite of these advances, we have yet to develop therapies based on the specific molecular defects present in this disease.
Claudin proteins are integral components of tight junctions and are often abnormally expressed in cancer.4, 5, 6 In particular, we and others have found that claudin-3, -4 and -7 are elevated in all subtypes of ovarian cancer.7, 8, 9, 10, 11, 12 Although the mechanisms and functional consequences of claudin overexpression remain unclear, the presence of these proteins on the surface of ovarian cancer cells suggests novel approaches for the therapy of this disease. Indeed, claudin-3 and -4 have been found to be natural receptors for Clostridium perfringens enterotoxin (CPE), a single chain 319 amino-acid toxin that leads to rapid cytolysis of cells expressing these proteins.13 CPE has therefore been proposed as a possible therapeutic agent in cancer cells that express high levels of claudin-3 and -4.4, 5, 14, 15, 16 Pre-clinical studies have demonstrated the efficacy of CPE in mouse xenograft models of ovarian, pancreatic, prostate and breast cancer.14, 17, 18, 19, 20
Although these studies are promising, the exact mechanism of action of CPE on cancer cells has not been clearly elucidated. In particular, how the presence of various combinations of claudins in the target cells may affect CPE efficacy is unclear. Here, we investigate the effect of CPE on multiple ovarian cancer cell lines that express different claudins. Interestingly, we find that cancer cells lacking claudin-3 and claudin-4 can still be sensitive to CPE and we identify for the first time claudin-6 as a functional receptor for CPE. A better understanding of CPE mechanism of action and, in particular, the identification of a previously unidentified functional receptor for CPE, may have important consequences as we continue to investigate CPE as a possible novel agent in cancer therapy.
Results
Claudin-3 and claudin-4 expression is not necessary for CPE action
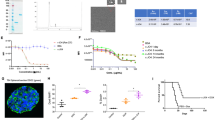
In order to study the effects of CPE on ovarian cancer cell survival, ovarian cancer cell lines Hey, A2780, BG1 and UCI-101 were treated with 0.1 or 1 μg/ml CPE for 2, 5, 15, 30 and 60 min, and then allowed to form colonies for 7 days (Figure 1a). The cell lines Hey and A2780 were not significantly affected by CPE treatment (0.1 and 1 μg/ml), regardless of treatment duration (Figures 1a and b). This is consistent with the fact that these lines do not express either of the two known CPE receptors (claudin-3 and claudin-4). On the other hand, cell line BG1, which expresses both receptors, was found to be sensitive to CPE and this effect was particularly obvious at 1 μg/ml (Figures 1a and b). Interestingly, cell line UCI-101, which does not express either receptor, was extremely sensitive to CPE, with significant decreases in the number of colonies even at the lower dose of CPE. Moreover, while treatment with the higher dose of CPE (1 μg/ml) shows equivalent sensitivity for both UCI-101 and BG1, a lower dose (0.1 μg/ml) of CPE allowed us to tease apart CPE sensitivity in these lines, with UCI-101 being significantly more sensitive than BG1, despite the absence of claudin-3 and -4 expression in UCI-101.
Effect of CPE on the viability and survival of ovarian cancer cell lines. (a) Ovarian cancer cell lines Hey, A2780, BG1 and UCI-101 were treated with 0.1 or 1 μg/ml CPE for 2, 5, 15, 30 and 60 min as indicated, and allowed to form colonies for a week before staining with crystal violet. Stained plates are shown for each condition. Top rows from each dish represent colonies from cells plated at 750 cells/well (low), while the bottom row shows colonies from cells plated at 3000 cells/well (high). (b) Quantitation of colony formation experiments from (a). Survival is expressed as percentage of surviving colonies compared with non-treated control for each cell line. Data are the means±s.e. of three independent experiments. An asterisk indicates a statistically significant difference (P<0.05).
Claudin-6 and -9 are highly homologous to claudin-3 and -4 and are expressed in ovarian cancer
As in vitro assays have shown that CPE can bind claudins other than claudin-3 and -4,21 we hypothesized that another claudin family member may be responsible for CPE cytotoxicity in UCI-101 cells. We performed a phylogenetic analysis of the second extracellular loop (ECL2) of claudin proteins, the known CPE-binding region in claudin-3 and -4 (Fujita et al.22 and Ling et al.23) and found that claudin-6 and -9 were the most closely related to claudin-3 and -4 in that region (Figure 2a). Indeed, 20 of 23 residues of the ECL2 were conserved in 3 of the 4 claudin sequences compared, including the NPLVA region previously reported important for CPE binding21 (Figure 2b). Interestingly, claudin-6 and -9 were found expressed in all four ovarian cancer cell lines examined (Figure 2c), while claudin-3 and -4 were only found to be expressed in BG1.
Claudin-6 and -9 are most closely related to claudin-3 and -4 (known CPE receptors). (a) Phylogenetic analysis of the ECL2 regions of classical claudins. Claudin-6 and -9 are most closely related to claudin-3 and -4, the known CPE receptors. (b) The sequence of the CPE-binding region is shown for claudin-3, -4, -6 and -9. (c) Immunoblot experiment showing the expression of claudin-3, -4, -6 and -9 in ovarian cancer cell lines. GAPDH was run as a loading control.
Expression of claudin-3, -4 or -6 can sensitize ovarian cell lines to CPE
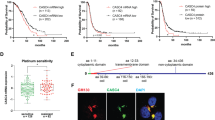
To test whether claudin-6 or -9 may act as a CPE receptor in ovarian cancer cells, we transiently transfected CPE-resistant ovarian cancer cell line Hey with claudin expression vectors for claudin-1, -3, -4, -6, -7, -9 or -14. These cells were also co-transfected with a green fluorescent protein (GFP) expression vector and, 48 h post transfection, treated with 1 μg/ml CPE. Upon CPE treatment, Hey cells transfected with claudin-3, -4 or -6 exhibited a significant decrease in GFP-positive cells (Figure 3a), indicating a sensitization of these cells to CPE. This decrease in GFP-positive cells was not seen in cells transfected with claudin-1, -7, -9 or -14. The same experiment was performed with another CPE-resistant ovarian cancer cell line, A2780, and identical results were obtained (Figure 3b), again demonstrating claudin-6 as a functional receptor for CPE cytotoxicity. In these experiments, transfection with claudin-6 did not lead to increased expression of claudin-3 or claudin-4 (data not shown).
Overexpression of claudin-3, -4 or -6 sensitizes CPE-resistant cell lines to CPE. Hey (a) or A2780 (b) cells were transfected with claudin-1, -3, -4, -6, -7, -9 or -14. GFP was co-transfected as a marker. The upper panel of images in each set represents a phase contrast images of cells and the lower panels are the fluorescent pictures of GFP-expressing cells for the same field. The graph on the right represents the number of fluorescent cells per field expressed as a percentage of their respective bright field controls. The data are representative of n=3 for claudin-3 and -4 and n=2 for claudin-6. Asterisks represent a statistically significant difference from control untreated cells with a P-value<0.05.
Knockdown of claudin-6 induces CPE resistance
Having established that overexpression of claudin-6 in CPE-resistant cell lines can sensitize them to CPE, we wondered whether the high sensitivity of UCI-101 to CPE was indeed due to claudin-6. To test this hypothesis, claudin-6 expression was knocked down in UCI-101 cells and these cells were then treated with 1 μg/ml CPE (Figure 4a). CPE treatment abolished colony formation, indicating that these cells are highly sensitive to CPE. Following claudin-6 knockout, however, several colonies were observed, indicating an important role for claudin-6 in CPE-mediated cytotoxicity in these cells. Identical results were obtained in OV90-P7, another claudin-6-expressing, CPE-sensitive cell line (Figure 4b) indicating that claudin-6-mediated CPE killing is not a rare event restricted to UCI-101 cells. The results from a similar experiment using claudin-6 small interfering RNA (siRNA) directed against a different part of the gene showed identical results (Supplementary Figure 1).
Knockdown of claudin-6 makes CPE-sensitive cell lines, UCI-101 and OV90-P7 resistant to CPE. UCI-101 (a) or OV90-P7 (b) cells were transfected with claudin-6 siRNA, treated with 1 μg/ml CPE 72 h post transfection, and grown for 7 days to allow colony formation. Claudin-6 knockdown reduced CPE effectiveness in both lines. The graphs represent the quantitation of these experiments (n=3 for both cell lines).
CPE can bind to claudin-6 in ovarian cancer cells
As claudin-6 can mediate CPE cytotoxicity in ovarian cancer cells, we wished to determine whether CPE could bind claudin-6 directly in these cells. Using a pull-down assay, we first confirmed that under our conditions, CPE binds to claudin-3 and -4 in the CPE-sensitive cell line BG1 (Figure 5a). Interestingly, CPE could bind to claudin-6 in the CPE-sensitive cell lines UCI-101 and BG1, but not in the CPE-resistant lines A2780 and Hey, although these cells do express claudin-6 (Figure 5b). CPE did not bind claudin-9 in any of the lines. Forced overexpression of claudin-6 in Hey cells, which sensitizes these cells to CPE toxicity (Figure 3a) led to binding of CPE to claudin-6 (Figure 5c).
CPE binds to claudin-6 in ovarian cancer cell lines. (a) Binding assays for CPE and claudin-3 and -4 in BG1 cells. CPE binds claudin-3 and -4 in this cell line. (b) claudin-6 and -9 binding to CPE in UCI-101, BG1, Hey and A2780 cell lines. CPE can bind claudin-6 in BG1 and UCI-101 cells. (c) Binding assay following claudin-6 transfection in the Hey ovarian cancer cell line. Although CPE does not bind endogenous claudin-6, overexpression of claudin-6 leads to binding of CPE to claudin-6.
The effect of CPE on a 3D spheroid model of ovarian cancer
To examine whether claudin-6 can also function as a functional CPE receptor in a spheroid 3D model, we created spheroid cultures of GFP-expressing A2780 (CPE resistant) and UCI-101 (claudin-6-dependent CPE sensitive). Treatment of the A2780 spheroids with CPE did not cause any change in their morphology or GFP fluorescence, while treatment of UCI-101 spheroids showed a decrease in the number of GFP-expressing cells in the spheroid, indicative of cell death (Figure 6).
The effect of CPE on 3D spheroid models of ovarian cancer. Ovarian cancer cell lines A2780 (a) and UCI-101 (b) stably transfected to express GFP were seeded at 2000, cells/well on matrigel and allowed to form spheroids. The spheroids were then treated with 1 μg/ml CPE for 24 h. Phase contrast and fluorescent images were taken on a Zeiss microscope at a magnification of × 10 24 h post treatment.
Discussion
Claudin-3 and -4 have been shown to be overexpressed in many epithelial tumors, including ovarian cancer.4, 5, 6 Interestingly, these claudin proteins may represent attractive therapeutic targets as they are the known receptors for the cytotoxic bacterial toxin CPE.4, 13, 14, 16 The potential anti-tumor effects of CPE have been studied in many different tumor types, including ovarian and uterine cancer.14, 24 Importantly, tumors resistant to conventional chemotherapy have been shown to be sensitive to CPE therapy provided they express claudin-3 or claudin-4.14 Various experiments in cell lines and mouse models have shown promising results for prostate, pancreatic, breast cancer and others, again provided that claudin-3 or -4 are expressed.4, 5, 16 Here, we show that the expression of claudin-3 or claudin-4 does not constitute an absolute requirement for CPE-mediated toxicity, and that claudin-6 can act as a functional receptor for CPE in ovarian cancer cells. This conclusion is based on several lines of evidence. (1) We show that claudin-6 overexpression in CPE-resistant cell lines is sufficient to sensitize these cells to CPE action (Figure 3). (2) claudin-6 knockdown in UCI-101 or OV90-P7, two CPE-sensitive lines that do not express claudin-3 or -4, can reduce CPE effectiveness (Figure 4). (3) These changes were accompanied by corresponding and consistent changes in claudin-6/CPE interactions (Figure 5). Our data therefore suggest that claudin-6 expression may represent an additional target/receptor for CPE cytotoxicity.
Unlike claudin-3 and -4, which are widely expressed in epithelial tissues, the expression of claudin-6 is more restricted and believed to be predominately found in embryonic tissues25 and in undifferentiated pluripotent stem cells.26 It has previously been reported that claudin-6 has an important role in the development of the mouse embryonic epithelium27 and endodermal tissues.28 However, its presence has been reported in some cancers such as rhabdoid tumors,29, 30 some ER-positive breast cancers,31 and gastric cancers.32 Our own findings show that claudin-6 can be expressed in ovarian cancer. The exact role of claudin-6 in cancer is unclear as its expression has been associated with both increased33 and decreased cancer aggressiveness.34 In any case, our findings suggest that CPE treatment may have a broader application in cancer treatment than initially thought.
Our binding assays (Figure 5) show that in CPE-sensitive UCI-101 cells CPE binds to claudin-6 (these cells do not express claudin-3 or -4). However, in CPE-resistant cell lines, Hey and A2780, which also expresses claudin-6, CPE does not bind claudin-6 or have cytotoxic effects unless claudin-6 is overexpressed by transient transfection. Therefore, the mere presence of claudin-6 is not sufficient for CPE binding and cytotoxic effects. CPE binding and effects may require a certain threshold of claudin-6 expression or a specific conformation, which may be affected by other TJ components. The effect does not seem related to claudin-6 localization, as Hey and A2780 cells express endogenous claudin-6 at the membrane (data not shown), but this claudin is somehow unavailable to bind CPE. However, transfection of claudin-6 in these cells led to the formation of a claudin-6-CPE complex and increased cell death. The exact mechanisms responsible for claudin-6 responsiveness to CPE treatment are currently under investigation and may involve claudin-6 phosphorylation (data not shown).
In vitro binding assays using short peptides have previously shown that CPE can bind claudin-6.21 In these assays, claudin-3, -6, -7, -9 and -14 were found to interact with CPE, although the functional significance of this binding, especially for claudin-6, -7 and -14 has remained unclear. Our finding that claudin-6 has a functional role in CPE action in cells shows that the previously reported in vitro binding can be functionally significant. Interestingly, these in vitro experiments failed to identify an interaction between CPE and claudin-4, suggesting that the 3D structure of the whole protein as well as possible interactions with other proteins may have an important role in CPE effects.
CPE is thought to induce cell lysis through a multistep process. Initially, CPE binds directly to claudin-3 and -4 to form a small 90 kDa SDS-sensitive complex (usually containing CPE, claudin-3/4 and claudin-1), which then oligomerizes/hexamerizes to form a much bigger complex and initiates CPE cytotoxicity.35 Our findings show that claudin-6 is an additional direct target of CPE (in addition to claudin-3/4) and would therefore be part of the initial small complex. We are currently investigating this model.
Although two-dimensional tissue culture-based assays are a routinely used for studying the efficacy of anti-tumor therapeutic candidates, this model does not accurately recapitulate the in vivo environment. Three-dimensional (3D) systems such as multicellular tumor spheroids better reflect the behavior of cells in tumors and, interestingly, claudins have been implicated in spheroid formation.36 We therefore used a 3D spheroid ovarian cancer model to test whether claudin-6 could also be used as a receptor under these conditions. Interestingly, multicellular tumor spheroid of UCI-101 cells (which express claudin-6, but not claudin-3 or -4) were sensitive to CPE, demonstrating that spheroid formation did not impede the ability of CPE to interact with claudin-6 and induce cell death.
It is clear that personalized molecular profiling will have an important role in the future of cancer therapy.37 Attacking specific pathway or targets known to be present in a particular cancer will dramatically improve the chance of efficient therapy. Although claudins are expressed in normal tissues, it may be possible to engineer a CPE derivative that specifically binds cancer-related claudins, which typically are not involved in forming tight junctions. In addition, our finding that claudin-6, a claudin not widely expressed in adult tissues but expressed in some cancers, is also a target for CPE cytotoxicity further expands the potential for CPE in cancer treatment.
Materials and methods
Cell lines and cell culture
The ovarian cancer cell lines Hey, BG1, A2780 and UCI-101 were maintained in McCoy’s complete medium (Gibco, Carlsbad, CA, USA) containing antibiotics (penicillin 100 U/ml and streptomycin 1000 μg/ml) (Gibco) and supplemented with 10% fetal bovine serum (Gibco). The OV90-P7 cell line was maintained in a 50:50 mixture of MCDB105 (Invitrogen, Carlsbad, CA, USA) and medium 199 supplemented with 15% fetal bovine serum (Gibco) containing antibiotics (penicillin 100 U/ml and streptomycin 1000 μg/ml). Cells were maintained in a 95% air and 5% CO2 humidified atmosphere at 37 °C. Cells were serially passaged when 75–80% confluent by trypsinization (0.25% 1X Trypsin without EDTA, Gibco) and resuspended in fresh medium.
Production of C. perfringens enterotoxin
The Histidine-tagged CPE protein in pET-16b expression vector (pET-HisCPE) has been published.19 A 10 ml culture of E. coli containing pET-10XHisCPE was grown overnight in LB with 1% glucose and 50 μg/ml ampicillin. One milliliter of the overnight culture was added to a 100-ml LB and allowed to grow at 37 °C to an absorbance of 0.3–0.4 at 600 nm, induced with 1 mM isopropyl-β-D-1 thiogalactopyranoside (IPTG)(Sigma-Aldrich, St Louis, MO, USA), and then allowed to grow overnight at 30 °C. The bacteria was then centrifuged at 5000 g for 20 min at 4 °C to pellet the cells and lysed in PBS with 1 mg/ml lyzozyme, protease and phosphatase inhibitors (Sigma-Aldrich), 1 mM phenylmethanesulfonylfluoride (Sigma-Aldrich) and 1 mg/ml DNAse I (Sigma-Aldrich). Sarcosine (Sigma-Aldrich) was added to a final concentration of 1.5% before sonication. The supernatant was then loaded on to a Ni-NTA agarose column (Qiagen, Germantown, MD, USA), washed twice with a Tris-HCl buffer (500 mM NaCl, 100 mM Tris-Hcl (pH 8.0)), followed by two washes in ice-cold PBS with 1% Triton-X 114 followed by elution with 200 mM Imidazole, dialyzed against PBS overnight at 4 °C and then sterile filtered.
Binding assays
The ovarian cancer cell lines Hey, BG1, UCI-101 and A2780 were seeded in 100 mm dishes at a density of 1 × 106 cells/dish. After 24 h the cells were collected with PBS containing protease and phosphatase inhibitors (Sigma-Aldrich) as well as 1% Triton-X 100 and CPE was added to the lysates for 30 min at 34 °C with constant gentle agitation. These lysates were then added to a Cu-chelate column for 3–4 h at 4 °C. The eluates were collected according to manufacturer’s protocol (Pierce, Pittsburgh, PA, USA) boiled and run on a 10% Tris-Glycine gel (Invitrogen). Proteins were analyzed by SDS–PAGE electrophoresis. Monoclonal mouse antibodies to the His-tag (Santa Cruz Biotechnology, Santa Cruz, CA, USA) and claudin-4 (Invitrogen), polyclonal rabbit antibodies to claudin-3 (Abcam, Cambridge, MA, USA), and claudin-9 (Novus Biologicals, Littleton, CO, USA) as well as a polyclonal goat antibody to claudin-6 (Santa Cruz Biotechnology) were used to detect proteins bound to CPE in the lysates. The membranes were then probed with the appropriate horseradish peroxidase-linked secondary antibody (Amersham Biosciences, Pittsburgh, PA, USA). Secondary antibodies were visualized by enhanced chemiluminescence western blotting detection reagents (Amersham).
Survival assays
The Hey, UCI-101, A2780 and BG1 ovarian cancer cell lines were seeded at a density of 750 or 3000 cells/well of a six-well dish. The following day cells were treated with 0.1μg/ml CPE for an hour. After washing out CPE, new medium was added to the cells and they were allowed to grow for 7 days following which the cells were stained with crystal violet and colonies were counted and quantified using NIH ImageJ software (http://rsbweb.nih.gov/ij/). For the survival assays following siRNA transfection, the cells were trypsinized 72 h post transfection, counted and reseeded at a density of 3000 cells/well. The colonies were allowed to grow for 7 days, stained with crystal violet and then quantified as described above.
Transfection
Hey, A2780 and UCI-101 cells were seeded in six-well tissue culture dishes at a density of 1.5 × 105 cells per dish in antibiotic-free medium. The following day, the Hey and A2780 cells were co-transfected with claudin- 1, -3, -4, -6, -7, -9 or -14 (2 μg each) along with GFP (0.5 μg) using Lipofectamine 2000 (Invitrogen) in optimem medium (Gibco) for 5–6 h. The transfection mixture was then aspirated and replaced with regular medium. The siRNA transfections in UCI-101 and OV90-P6 were carried out in a similar manner with both the control and claudin-6 siRNA being used at a concentration of 100 nM per transfection.
Immunoblotting
Cells were washed with Hanks buffered saline solution and cell lysates were prepared in lysis buffer (150 mM Tris-HCl, pH 6.8, 25% glycerol and 5% SDS). The proteins from the cell lysates were separated by SDS–PAGE in 4–12 or 10–20% Tris-Glycine gels and transferred to PVDF membranes (Millipore, Billerica, MA, USA). The membranes were blocked for 1 h at RT with TBS-T+5% non-fat dry milk (Carnation) and incubated overnight at 4 °C with antibodies specific for the indicated proteins (monoclonal mouse antibodies to the His-tag (Santa Cruz Biotechnology), GAPDH (Abcam)) and β-actin (Abcam) and claudin-4 (Invitrogen), polyclonal rabbit antibodies to claudin-3 (Abcam) and claudin-9 (Novus Biologicals) as well as a polyclonal goat antibody to claudin-6 (Santa Cruz Biotechnology). After washing the membranes thrice with TBS-T (TBS with 1% Tween-20), they were incubated with horseradish-peroxidase conjugated secondary antibodies and detected by enhanced chemiluminescence(Amersham Biosciences).
Generation of GFP-expressing stable transfectants
UCI-101 and A2780 cells were seeded in six-well dishes at a density of 1.5 × 105 cells per dish in antibiotic-free medium. The following day, they were transfected with 1.0 μg of a GFP-expressing plasmid containing a selection marker using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s protocol. The cells were then selected for growth in Geneticin-containing medium and sequentially cloned to produce stable GFP-expressing cell lines.
Spheroid model of ovarian cancer
Forty-eight-well tissue culture dishes (Mattek, Ashland, MA, USA) were refrigerated for an hour before adding 200 μl of matrigel (BD Biosciences, San Jose, CA, USA) to each well. These dishes were then incubated at 37 °C for 10 min before plating. The UCI-101 and A2780 cell lines generated to stably express GFP were seeded at 2000, cells/well in medium to which 5% matrigel was added, changing the medium every 48 h. Cells were treated with 1 μg/ml CPE for an hour. Spheroids were visualized with a Zeiss microscope (Thornwood, NY, USA) using AxioRel 7.0 software (Carl Zeiss). Loss of viability due to treatment with CPE was measured by a decrease in GFP signal.
Statistical analysis
GraphPad Prism version 3.0 (San Diego, CA, USA) was used to perform statistical analysis among different experimental groups. One-way analysis of variance with a Bonferroni post-test was used when performing multiple sample comparisons, whereas a two-tailed, unpaired t-test was performed when comparing two experimental groups. The results are presented as mean±s.e. of the mean or s.d. of the mean for all experiments, and for all statistical analysis, P<0.05 was considered statistically significant unless stated otherwise.
References
Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D . Global cancer statistics. CA Cancer J Clin 2011; 61: 69–90.
Bast RC, Hennessy B, Mills GB . The biology of ovarian cancer: new opportunities for translation. Nat Rev Cancer 2009; 9: 415–428.
Hennessy BT, Coleman RL, Markman M . Ovarian cancer. Lancet 2009; 374: 1371–1382.
Morin PJ . Claudin proteins in human cancer: promising new targets for diagnosis and therapy. Cancer Res 2005; 65: 9603–9606.
Kominsky SL . Claudins: emerging targets for cancer therapy. Expert Rev Mol Med 2006; 8: 1–11.
Valle B, Morin PJ . Claudins in cancer biology. Curr Top Membr 2010; 13: 293–333.
Hough CD, Sherman-Baust CA, Pizer ES, Montz FJ, Im DD, Rosenshein NB et al. Large-scale serial analysis of gene expression reveals genes differentially expressed in ovarian cancer. Cancer Res 2000; 60: 6281–6287.
Rangel LBA, Agarwal R, D’Souza T, Pizer ES, Alò PL, Lancaster WD et al. Tight junction proteins claudin-3 and claudin-4 are frequently overexpressed in ovarian cancer but not in ovarian cystadenomas. Clin Cancer Res 2003; 9: 2567–2575.
Santin AD, Zhan F, Bellone S, Palmieri M, Cane S, Bignotti E et al. Gene expression profiles in primary ovarian serous papillary tumors and normal ovarian epithelium: identification of candidate molecular markers for ovarian cancer diagnosis and therapy. Int J Cancer 2004; 112: 14–25.
Bignotti E, Tassi RA, Calza S, Ravaggi A, Romani C, Rossi E et al. Differential gene expression profiles between tumor biopsies and short-term primary cultures of ovarian serous carcinomas: identification of novel molecular biomarkers for early diagnosis and therapy. Gynecol Oncol 2006; 103: 405–416.
Kleinberg L, Holth A, Fridman E, Schwartz I, Shih Ie M, Davidson B . The diagnostic role of claudins in serous effusions. Am J Clin Pathol 2007; 127: 928–937.
Dahiya N, Becker KG, Wood WH, Zhang Y, Morin PJ . Claudin-7 is frequently overexpressed in ovarian cancer and promotes invasion. PLoS ONE 2011; 6: e22119.
Katahira J, Sugiyama H, Inoue N, Horiguchi Y, Matsuda M, Sugimoto N . Clostridium perfringens enterotoxin utilizes two structurally related membrane proteins as functional receptors in vivo. J Biol Chem 1997; 272: 26652–26658.
Santin AD, Cane S, Bellone S, Palmieri M, Siegel ER, Thomas M et al. Treatment of chemotherapy-resistant human ovarian cancer xenografts in C.B-17/SCID mice by intraperitoneal administration of Clostridium perfringens enterotoxin. Cancer Res 2005; 65: 4334–4342.
Takahashi A, Kondoh M, Suzuki H, Yagi K . Claudin as a target for drug development. Curr Med Chem 2011; 18: 1861–1865.
Walther W, Petkov S, Kuvardina ON, Aumann J, Kobelt D, Fichtner I et al. Novel Clostridium perfringens enterotoxin suicide gene therapy for selective treatment of claudin-3- and -4-overexpressing tumors. Gene Ther 2012; 19: 494–503.
Michl P, Buchholz M, Rolke M, Kunsch S, Lohr M, McClane B et al. Claudin-4: a new target for pancreatic cancer treatment using Clostridium perfringens enterotoxin. Gastroenterology 2001; 121: 678–684.
Kominsky SL, Vali M, Korz D, Gabig TG, Weitzman SA, Argani P et al. Clostridium perfringens enterotoxin elicits rapid and specific cytolysis of breast carcinoma cells mediated through tight junction proteins claudin 3 and 4. Am J Pathol 2004; 164: 1627–1633.
Santin AD, Bellone S, Siegel ER, McKenney JK, Thomas M, Roman JJ et al. Overexpression of Clostridium perfringens enterotoxin receptors claudin-3 and claudin-4 in uterine carcinosarcomas. Clin Cancer Res 2007; 13: 3339–3346.
Maeda T, Murata M, Chiba H, Takasawa A, Tanaka S, Kojima T et al. Claudin-4-targeted therapy using Clostridium perfringens enterotoxin for prostate cancer. Prostate 2012; 72: 351–360.
Winkler L, Gehring C, Wenzel A, Muller SL, Piehl C, Krause G et al. Molecular determinants of the interaction between Clostridium perfringens enterotoxin fragments and claudin-3. J Biol Chem 2009; 284: 18863–18872.
Fujita K, Katahira J, Horiguchi Y, Sonoda N, Furuse M, Tsukita S . Clostridium perfringens enterotoxin binds to the second extracellular loop of claudin-3, a tight junction integral membrane protein. FEBS Lett 2000; 476: 258–261.
Ling J, Liao H, Clark R, Wong MS, Lo DD . Structural constraints for the binding of short peptides to claudin-4 revealed by surface plasmon resonance. J Biol Chem 2008; 283: 30585–30595.
Santin AD, Bellone S, Marizzoni M, Palmieri M, Siegel ER, McKenney JK et al. Overexpression of claudin-3 and claudin-4 receptors in uterine serous papillary carcinoma: novel targets for a type-specific therapy using Clostridium perfringens enterotoxin (CPE). Cancer 2007; 109: 1312–1322.
Hewitt KJ, Agarwal R, Morin PJ . The claudin gene family: expression in normal and neoplastic tissues. BMC Cancer 2006; 6: 186.
Wang L, Xue Y, Shen Y, Li W, Cheng Y, Yan X et al. Claudin 6: a novel surface marker for characterizing mouse pluripotent stem cells. Cell Res 2012; 22: 1082–1085.
Turksen K, Troy TC . Claudin-6: a novel tight junction molecule is developmentally regulated in mouse embryonic epithelium. Dev Dyn 2001; 222: 292–300.
Anderson WJ, Zhou Q, Alcalde V, Kaneko OF, Blank LJ, Sherwood RI et al. Genetic targeting of the endoderm with claudin-6CreER. Dev Dyn 2008; 237: 504–512.
Birks DK, Kleinschmidt-DeMasters BK, Donson AM, Barton VN, McNatt SA, Foreman NK et al. Claudin 6 is a positive marker for atypical teratoid/rhabdoid tumors. Brain Pathol 2010; 20: 140–150.
Sullivan LM, Yankovich T, Le P, Martinez D, Santi M, Biegel JA et al. Claudin-6 is a nonspecific marker for malignant rhabdoid and other pediatric tumors. Am J Surg Pathol 2012; 36: 73–80.
Yafang L, Qiong W, Yue R, Xiaoming X, Lina Y, Mingzi Z et al. Role of estrogen receptor-alpha in the regulation of claudin-6 expression in breast cancer cells. J Breast Cancer 2011; 14: 20–27.
Rendon-Huerta E, Teresa F, Teresa GM, Xochitl GS, Georgina AF, Veronica ZZ et al. Distribution and expression pattern of claudins 6, 7, and 9 in diffuse- and intestinal-type gastric adenocarcinomas. J Gastrointest Cancer 2010; 41: 52–59.
Zavala-Zendejas VE, Torres-Martinez AC, Salas-Morales B, Fortoul TI, Montano LF, Rendon-Huerta EP . Claudin-6, 7, or 9 overexpression in the human gastric adenocarcinoma cell line AGS increases its invasiveness, migration, and proliferation rate. Cancer Invest 2011; 29: 1–11.
Wu Q, Liu Y, Ren Y, Xu X, Yu L, Li Y et al. Tight junction protein, claudin-6, downregulates the malignant phenotype of breast carcinoma. Eur J Cancer Prev 2010; 19: 186–194.
Mitchell LA, Koval M . Specificity of Interaction between Clostridium perfringens enterotoxin and Claudin-Family tight junction proteins. Toxins 2010; 2: 1595–1611.
Boylan KL, Misemer B, Derycke MS, Andersen JD, Harrington KM, Kalloger SE et al. Claudin 4 is differentially expressed between ovarian cancer subtypes and plays a role in spheroid formation. Int J Mol Sci 2011; 12: 1334–1358.
Workman P, Clarke PA, Al-Lazikani B . Personalized medicine: patient-predictive panel power. Cancer Cell 2012; 21: 455–458.
Acknowledgements
This research was supported in part by the Intramural Research Program of the NIH, National Institute on Aging. ADS is Supported in part by grants from NIH R01 CA122728-01A4 and R01 CA154460-01A1, the Honorable Tina Brozman Foundation and Deborah Bunn Alley Ovarian Cancer Research Foundation. We thank the members of our laboratory for helpful comments on the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on the Oncogenesis website .
Supplementary information
Rights and permissions
This work is licensed under the Creative Commons Attribution-NonCommercial-No Derivative Works 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/3.0/
About this article
Cite this article
Lal-Nag, M., Battis, M., Santin, A. et al. Claudin-6: a novel receptor for CPE-mediated cytotoxicity in ovarian cancer. Oncogenesis 1, e33 (2012). https://doi.org/10.1038/oncsis.2012.32
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/oncsis.2012.32
Keywords
This article is cited by
-
The role of claudins in cancer metastasis
Oncogene (2017)
-
Claudins in cancer: bench to bedside
Pflügers Archiv - European Journal of Physiology (2017)
-
Pore-forming toxins: ancient, but never really out of fashion
Nature Reviews Microbiology (2016)