Abstract
ΔNp63α is a critical mediator of epithelial development and stem cell function in a variety of tissues including the skin and breast, while overexpression of ΔNp63α acts as an oncogene to drive tumor formation and cancer stem cell properties in squamous cell carcinoma. However, with regards to the prostate, while ΔNp63α is expressed in the basal stem cells of the mature gland, during adenocarcinoma development, its expression is lost and its absence is used to clinically diagnose the malignant state. Surprisingly, here we identify a sub-population of bone metastatic prostate cancer cells in the PC3 cell line that express ΔNp63α. Interestingly, we discovered that ΔNp63α favors adhesion and stem-like growth of these cells in the bone microenvironment. In addition, we show that these properties require expression of the target gene CD82. Together, this work uncovers a population of bone metastatic prostate cancer cells that express ΔNp63α, and provides important information about the mechanisms of bone metastatic colonization. Finally, we identify metastasis-promoting properties for the tetraspanin family member CD82.
Similar content being viewed by others
Introduction
P63 is a member of the p53 family of transcription factors, and contains two main isoforms of the protein, designated transactivating and deltaN (ΔN). Further variability is given by alternative splicing, generating three main variants for each isoform (α, β and γ).1 In general, the TAp63 isoforms function more similarly to p53 in regulating apoptosis and senescence, while ΔNp63 has been linked with stem cell fate and proliferation.2, 3
In tumor development, ΔNp63α is overexpressed in the majority of human squamous cell carcinoma, and recent studies demonstrated an oncogenic role for this isoform during squamous cell carcinoma formation.4, 5 With regards to prostate cancer (PC), however, there is a different expression pattern. P63 is detected in the basal stem cells of the normal tissue, and is suggested to identify the tumor-initiating population in mouse and human cancers.6, 7 Subsequently the expression of ΔNp63α is lost during the transformation process and growth of the primary prostate tumor. As such, PC is negative for ΔNp63α, and assessment of p63 negativity is used clinically to diagnose tumor status.3, 7, 8, 9, 10
With the aim of modeling metastatic PC, many studies have benefited from the use of three cell lines derived from individual metastatic sites, to investigate the cellular and molecular processes involved. These include PC3 cells, derived from bone metastasis, DU145 cells from brain and LNCaP from lymph node. Within these, only PC3 is capable of forming bone metastasis in mouse when the cells are introduced via intra-cardiac or intra-tibial injection. Interestingly, this cell line contains cancer stem-like cells that are more aggressive in forming tumors in vivo.11, 12, 13 In agreement with clinical data, all these cell lines have been described to be negative for p63 expression.
Unexpectedly we identified a sub-population of cells in the PC3 line that expresses ΔNp63α. Through functional misexpression studies in vivo, coupled with detailed molecular profiling, we discovered that ΔNp63α contributes to the adhesion and colonization of these bone metastatic PC cells. Interestingly, these cells have a shared signature with normal stem-like cells, and in particular, increased expression of the adhesion molecule CD82. Surprisingly, functional follow-up studies identified pro-metastatic functions for CD82, through the regulation of cell adhesion. Together, this identifies previously unknown heterogeneity in the PC3 bone metastatic cell line, and further supports how promotion of stem-like properties may favor metastatic growth.
Results
ΔNp63 expression is enriched in the basal stem cells of the adult mouse prostate
Studies with p63-deficient models have demonstrated a requirement for p63 in the development of the prostate.14, 15, 16 In addition, p63 expression has been documented in basal cells in the adult gland that express stem cell markers.7, 17 These observations suggest critical roles for p63 in adult prostate stem cells (PSC), yet the underlying mechanisms remain unknown. We validated previous studies demonstrating specific p63 expression in the small Keratin 5-positive basal cells of the prostate, and in close proximity to the Keratin 18 luminal cells, which are negative for p63 (Supplementary Figures 1A,B). Next, we dissociated the glands to a single cell suspension, and fluorescence-activated cell sorting (FACS) sorted the basal stem cell population using the previously characterized PSC markers CD49f and Sca-1.17, 18 By performing RT–qPCR, we showed that ΔNp63 expression is highest in the double-positive CD49f/Sca-1 stem cells (Supplementary Figure 1C, D). We confirmed this at the protein level, performing cytospin-immunostaining on the same populations (Supplementary Figures 1E and F).
ΔNp63α is expressed in a sub-population of human metastatic PC cells
Unlike the normal prostate gland, PC and PC cell lines are considered negative for p63 expression.8, 10, 19 Surprisingly however, we detected expression of ΔNp63 in PC3 cells, an aggressive cancer cell line derived from metastasis to the bone. Indeed, very low levels of the ΔNp63 transcript was only detectable in this cell line, and not in others derived from prostate metastasis to the lymph node (LNCaP) and brain (DU145) (Figure 1a). Interestingly, the expression of the transcript increased in high-density cultures (100% confluency) compared to low density/exponential phase proliferating cells (60–70% confluency). At high density, there was no increase in apoptosis levels (Supplementary Figure 2A), possibly reflecting a function for ΔNp63 in survival at high density through reversible growth inhibition and acetylation.20 Performing immunoprecipitation (IP) western blotting in PC3 cells grown at high density to enrich for possible low-level expression, we could detect ΔNp63α protein. As a control, we used RWPE-1 cells, a non-transformed line enriched in basal/stem cells, with high levels of the ΔNp63α (Figure 1b). Again, expression was not detectable in LNCaP or DU145.
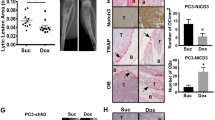
ΔNp63α is expressed in a sub-population of PC3 human bone metastatic cells. (a) RT–qPCR for the ΔN and transactivating isoforms of p63 in four prostate cell lines: RWPE-1, PC3, LNCaP and DU145. Low density (60–70% confluency), LD; high density (100% confluent), HD. Values are compared to those of RWPE-1 LD replicate as reference. Graphs show results as mean±s.e.m. of three biological replicates. (b) IP and western blot for p63 in the four prostate cell lines. Input, IN; IP with immunoglobulin G control, IgG; IP with the p63-specific antibody, IP. Arrows denote the height where the ΔNp63α band is expected and asterisks indicate unspecific bands in the LNCaP input lane that are not present in the IP lane. (c) Immunofluorescence for p63 on RWPE-1 and PC3 cells in culture. (d) PC3 cells were infected with a reporter-vector and injected intra-tibially in adult nude mice. The derived bone metastases were identified by immunohistochemical staining for GFP. P63 (and specifically ΔNp63 expression) was found in a sub-population of GFP+ metastatic cells in bone.
Given that ΔNp63α was present at such low levels in PC3, this suggested that expression might be low in every cell, or that only some cells might express detectable protein. To address this, we performed immunocytochemistry in culture. Remarkably, this identified that a sub-population of cells specifically expressed p63 in PC3 (Figure 1c) and not in the non-bone metastatic cell lines LNCaP and DU145 (Supplementary Figure 2B). However, as the PC3 line is derived from a bone metastatic tumor, we wanted to examine whether we could detect a similar pattern of p63 in bone metastatic prostate tumors in vivo. To achieve this, we performed intra-cardiac injections of the PC3 cell line, an assay to assess the potential homing and colonization to metastatic sites, as well as intra-tibial injections, which measures the ability of metastatic cells to adhere and grow within the bone microenvironment. These two models are particularly useful as there are no genetically engineered mouse models that spontaneously metastasize to the bone.21, 22 Subsequently, the tumors that developed were stained for total p63 and ΔNp63. As is shown in Figure 1d, intra-tibial tumors in the bone were stained with H&E and Massons Trichrome to identify the location of the bone. Then immunostaining for p63 (all isoforms) and the ΔNp63 isoform showed that, in agreement with our in vitro data, individual p63- and ΔNp63-positive cells were detectable in the metastatic lesions that developed in the bone (Figure 1d and Supplementary Figure 2C). This surprising observation uncovers previously unknown heterogeneity in the PC3 cell line.
ΔNp63α promotes colonization of prostate metastatic cells to the bone
Next, we investigated the effects of alteration of ΔNp63 expression on the PC3 cell line in vivo. First, we overexpressed ΔNp63α in PC3 cells, along with a luciferase vector, and directly injected 250 000 cells into the tibia of nude mice to measure for effects on tumor growth in the bone (Figure 2a). Here the effects were striking, in that enhanced expression of ΔNp63α in bone metastatic PC cells increased the early colonization and growth of the bone metastatic lesions in vivo. For example, at 3 weeks post-injection, 5/7 mice receiving control cells did not yet display any tumor-bearing luciferase signal, whereas 7/9 mice that had been injected with ΔNp63α, already had detectable tumor growth (Figure 2a). This increased tumor intensity persisted throughout the course of the study, resulting in the ΔNp63α-expressing mice having to be killed/dying sooner than controls (table inset, Figure 2a). These differences were less obvious however when higher numbers of cells (1 × 106) cells were injected (data not shown), further highlighting the role of p63 in early metastatic growth.
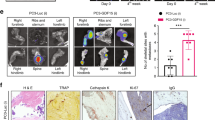
ΔNp63α confers a growth advantage and stem-like properties to PC3 cells in the bone microenvironment. (a) Adult male nude mice were injected intra-tibially with 2.5 × 105 PC3 cells infected with vector control (P-V) or overexpressing ΔNp63α (P-ΔN) and co-infected with the Luc-GFP construct. Bone metastasis was monitored weekly by luciferase imaging beginning from week 3. Seven mice were analyzed for the P-V group and nine for the P-ΔNα group. Animals were killed at different time points depending on the appearance of the tumor. The table indicates the total number of mice that were measured for a luciferase signal at each time point. Bone metastasis luciferase signal and tumor incidence are represented as previously specified and P-values are calculated using the log-rank test. (b) P-V and P-ΔNα were plated at the density of 100 cells/well in triplicate in six-well plates and after 2 weeks the colonies were stained with crystal violet. Colonies were counted and their morphology analyzed through light microscopy. Results are presented as mean±s.e.m. of three biological replicates.
Interestingly, the PC3 cell line contains a population of holoclone cells that display cancer stem-like properties and that contribute to tumor growth and renewal (Doherty et al.11, Li et al.12, Zhang and Waxman 13). Given that p63 is a key mediator of stem-like properties in normal prostate and other cancer types, we next asked whether increased expression of ΔNp63α had any effect on holoclone growth patterns in PC3 cells. Interestingly, upon overexpression of ΔNp63α, there was a significantly higher number of holoclones and fewer paraclones, supporting the idea that ΔNp63α might contribute to the stem-like properties in these cells (Figure 2b).
These findings suggest that an investigation of ΔNp63α in these cells might serve to uncover mechanistic links between stem-like properties and metastatic tumor growth. To address this, we first generated shRNAs specifically targeting the ΔNp63 isoform, and validated their specificity in the non-transformed prostate cell line RWPE-1, that has basal stem-like characteristics (Supplementary Figure 3A). Interestingly, knockdown of ΔNp63 in this cell line led to a decrease in growth ability as measured by growth curve and focus-formation assays (Supplementary Figures 3B,C). In addition, the RWPE-1 cell line, due to its stem-like properties, forms spheres in 3D culture. These features were also significantly impaired with knockdown of ΔNp63, resulting in fewer and smaller spheres at first and second passage, supporting a role for ΔNp63α in regulating adult PSC growth (Supplementary Figure 3D).
ΔNp63 directly controls expression of genes involved both in epithelial stem cell maintenance, bone survival and adhesion
As a first step to identify prostate-specific genes whose expression is regulated by ΔNp63α, we performed microarray analysis on the RWPE-1 cell line following knockdown of the ΔN isoform, which causes loss of stem-like proliferative capacity in these cells. We identified a total of 581 genes whose expression increased, and 676 genes whose expression decreased upon knockdown of ΔNp63 (Figure 3a). Interestingly, loss of ΔNp63 induced a downregulation of basal markers, and an increase in luminal markers (Figure 3b), as well as decreased expression of known stem cell genes, including WNT3A, CD44, ITGα6, NOTCH1 and FGFR2 (Figures 3b and c).
ΔNp63 directly regulates genes involved in epithelial development, stem cells and the bone microenvironment. (a) Microarray analysis of RWPE-1 cells infected with a short hairpin for ΔNp63 (R-shΔN) compared to RWPE-1 infected with vector control (R-V) was performed. The heat map represents the microarray results, with a fold change threshold of 1.4 and the P-value at 0.05. (b) Depiction of basal, luminal and stem cell genes that were up- and downregulated in R-shΔN, compared to R-V in the microarray. (c) RT–qPCR validation of some of the genes identified in the microarray. cDNA fold changes are compared to values in R-V. Two biological replicates are presented for ΔNp63 and TAp63, and one for the other genes. (d) ChIP-Seq analysis for p63 was performed in RWPE-1. Mapping peaks of p63 targets p21, FGFR2, WNT3A, BDKRB1 and MMP14 are shown. (e) Volcano plot of the common genes found in the microarray with ΔNp63 knockdown and ChIP-Seq. (f) Genomatix software analysis of direct p63-target genes downregulated in microarrays of R-shΔN compared to R-V showing the over-represented pathways, tissues and biological processes (full analysis and gene lists in Supplementary Table 2).
Next, we wanted to identify those genes that are direct transcriptional targets of p63 in prostate cells. While this has been done for keratinocytes, similar data for the prostate have not been generated. With this purpose, we performed chromatin-IP followed by high-throughput sequencing (ChIP-Seq) on RWPE-1 cells. Importantly, this identified a number of previously known targets of p63, including P21, PERP, CD82 and FGFR2, as well as new ones including WNT3A and MMP14 (Figure 3d). Then, by overlapping the complete list of p63 targets with our microrarray data set, we could identify those p63-target genes whose expression was altered by loss of ΔNp63, and that might mediate the stem-like properties of p63 in the prostate (Figure 3e). As ΔNp63α can act both as a transcriptional repressor or an activator of gene expression, we were initially interested in identifying those p63 targets whose expression was activated by p63, so we focused on the downregulated list. Using the Genomatix software to perform Gene Ontology and Pathway analysis, we identified interesting signatures (Figure 3f and Supplementary Table 1). As might be expected, in prostate cells, ΔNp63 expression correlated with pathways relating to epithelial status and stem cell function. Surprisingly however, terms relating to bone were also identified, including ‘osteoblasts’, ‘bone matrix’ and ‘osteocytes’, suggesting that, even in basal stem-like cells, ΔNp63α mediates a set of genes that are related to bone survival and growth. Moreover, among the most prominent signatures detected were ‘adhesion’ and adhesion-related events including ‘ECM’ and ‘basement membrane’, suggesting that ΔNp63 might be regulating these properties in PSCs.
To determine whether ΔNp63α might be regulating similar processes in the metastatic cancer cell line, we performed microarray on PC3 cells in which ΔNp63α was overexpressed, and which exhibit increased bone metastasis in vivo. This identified a total of 43 upregulated and 19 downregulated genes (Figure 4a). Then finally, by comparing those lists of genes that exhibited a significant increase in PC3 cells upon ΔNp63α overexpression, with the list of direct p63-target genes that displayed significantly reduced expression upon knockdown of ΔNp63α in RWPE-1 cells, we were able to identify 11 genes in common, that are directly activated by ΔNp63α in normal PSCs and PC bone metastasis (Figures 4b and c). Remarkably, these genes could be classified into functional categories, including ‘adhesion’ and ‘homing to the bone’, as well as ‘drug metabolism’ and ‘membrane transport’. Furthermore, those genes linked to adhesion and bone homing (CD82, ITGB4, IL1B and DST) have previously been linked with p63 expression in other settings,23, 24, 25, 26 supporting the specificity of our findings in PC.
ΔNp63 directs a signature of genes favoring stemness and adhesion in prostate metastasis. (a) Microarray analysis was performed on P-ΔNα, compared to P-V. The heat map represents results setting the fold change threshold at 1.4 and the P-value at 0.05. (b) Representational comparison of genes upregulated by overexpression of ΔNp63α in PC3, downregulated in RWPE-1 knocked down for ΔNp63 and that are direct targets of p63 identified by ChIP-Seq. (c) RT–qPCR for the validation of some direct targets of p63 changing accordingly with ΔNp63 both in PC3 and RWPE-1. Results presented as mean±s.e.m. from four and three biological replicates, respectively, for PC3 and RWPE-1. (d) P-V and P-ΔNα were plated on fibronectin- or laminin I-coated plates, and the number of cells adhering was counted. Results are presented as mean±s.e.m. of three technical replicates of one representative biological replicate of three attempts.
The p63-target CD82 mediates bone metastatic adhesion
Given that cellular adhesion emerged as a major molecular signature in our profiling studies, and the increasing link with p63 and adhesive function in different settings,27, 28, 29 we investigated if this might also be important in prostate bone metastatic cell behavior. First, we checked whether increased expression of ΔNp63α in PC3 cells might confer increased adhesive properties on different ECM substrata (Figure 4d). Importantly, overexpression of ΔNp63α caused a significant increase in adhesion in every condition tested, including fibronectin and laminin I (Figure 4d), as well as collagen I (not shown).
Of the adhesion-related genes identified in our analysis, CD82 (also known as KAI-1) is a well-known epithelial-mesenchymal transition (EMT) suppressor and has been linked with adhesion and homing of hematopoietic and leukemic stem cells to the bone microenvironment, prompting us to investigate it further.30, 31, 32 Interestingly, we found that PC3 cells contain a sub-population of CD82-positive cells, which, upon ΔNp63α overexpression was significantly increased (Figure 5a). Since CD82 expression was also only detected in some cells in a manner similar to ΔNp63, we investigated whether these might represent a similar sub-population in prostate metastatic cells. To address this, we purified the CD82-high cells from the PC3 population and examined for expression of the 11 ΔNp63-regulated genes (Figure 5b). Interestingly, many of these same genes were expressed at significantly higher levels in the CD82-high population, supporting the concept that CD82 might be a mediator of the adhesion and metastatic properties of ΔNp63.
The p63-target CD82 controls metastatic adhesion mediated by ΔNp63α in vitro and in vivo in the bone microenvironment. (a) PC3 cells infected with V or ΔNp63α were stained for CD82 and analyzed by FACS. Representative FACS plot and graph showing the percentage of CD82+ cells as mean±s.e.m. of three biological replicates. (b) CD82+ and CD82− cells were sorted from PC3. Levels of the two isoforms of p63 and of other genes directly regulated by ΔNp63 were analyzed by RT–qPCR in the two populations. One representative FACS plot of three experiments is shown and graph depicts results as mean±s.e.m. from three biological replicates, with values compared to the PC3 CD82− population. (c) shRNAs targeting CD82 were infected into PC3 cells overexpressing ΔNp63α. The efficiency of knockdown was tested by RT–qPCR (transcript) and FACS (protein). RT–qPCR values were compared to cells infected with a short hairpin control (shct) and results are presented as mean±s.e.m. of three biological replicates. Representative FACS plots depict one of three experiments. (d) P-ΔNα+shct, P-ΔNα+shCD82 1 and P-ΔNα+shCD82 2 were plated in the presence of fibronectin or laminin I. An adhesion assay was performed as previously described in Figure 4. Results are presented as mean±s.e.m. of three technical replicates of one representative biological replicate of two attempts. (e) P-ΔNα cells were treated with different concentrations of CD82-blocking antibody and plated in the presence of fibronectin. An adhesion assay was performed as previously described in Figure 4. Results are presented as mean±s.e.m. of three technical replicates of one biological replicate. (f) Adult male nude mice were injected intra-tibially with 2.5 × 105 P-ΔNα+shct+Luc-GFP and P-ΔNα+shCD82+Luc-GFP. Bone metastasis was monitored weekly by luciferase imaging starting from week 3. Ten mice were analyzed for the P-ΔNα+shct group and seven for the P-ΔNα+shCD82 group. Animals were killed at week 6. Imaging of mice at week 6 is shown and luciferase signal is represented as mean±s.e.m. The tumor incidence refers to the total number of mice developing lesions from the beginning of the study, P-values are calculated using the log-rank test.
To investigate this possibility, we obtained shRNAs targeting CD82, and tested if these hairpins could successfully disrupt expression at the transcript and protein level, in PC3 cells overexpressing ΔNp63α (Figure 5c). Next, using the two most effective shRNAs with high knockdown efficiency, we examined whether knockdown of CD82 might diminish the adhesion mediated by ΔNp63α. Indeed, in PC3 cells overexpressing ΔNp63α, knockdown of CD82 with two separate hairpins abolished the adhesion of these cells, identifying this gene as a key player in ΔNp63-mediated adhesion (Figure 5d). To further investigate the possibility that targeted intervention with CD82-mediated signaling might affect ΔNp63α-driven adhesion, we treated the cells with an antibody to CD82 and found that interference with the protein also significantly reduced adhesion in culture (Figure 5e). Together, this suggests that CD82 might be a novel regulator of ΔNp63α function in PC3 cells.
However, the most important test was to identify whether loss of CD82 also functioned to disrupt ΔNp63α-driven bone metastatic tumor growth in vivo. To address this, we again performed intra-tibial injections of the PC3-ΔNp63α cells, this time with or without co-expression of the CD82 hairpin (Figure 5d). As before, PC3 cells with overexpression of ΔNp63α formed large tumors that developed at early stages following injection. Remarkably however, in the absence of CD82, metastatic tumor growth was practically non-existent. Together, this demonstrates that CD82-mediated adhesion, regulated by ΔNp63α, directly contributes to bone metastatic tumor colonization and growth, and suggests possible new avenues for therapeutic intervention.
CD82 mediates adhesion in other metastatic PC cells
These results uncovered previously unknown heterogeneity in the bone metastatic PC3 cell line, identifying a sub-population of cells that express ΔNp63α, and which use CD82 to contribute to tumor cell adhesion and metastatic growth. This raises the question of whether these findings can provide information on broader mechanisms in epithelial tumor growth and metastasis. As described previously,33 CD82 has a wider expression in other PC metastatic cell lines (Figure 6a). To investigate if CD82 regulates a broader program of adhesion, we knocked down CD82 in these three main PC metastatic cell lines: PC3, DU145 and LNCaP (Figure 6b). Interestingly, knockdown of CD82 in each of the three cell lines led to changes in growth rates or proliferation, but the cells were still viable (Figure 6c). However, when subjected to adhesion assays, there was a clear and significant loss of adhesion in each cell type upon CD82 knockdown 5 (Figure 6d), suggesting that this protein may play previously unknown roles in promoting metastatic colonization.
CD82 mediates adhesion also of non-bone metastatic PC cells. (a) FACS plots of PC3, DU145 and LNCaP stained for CD82. The percentage of positive cells is indicated. (b) The shRNAs targeting CD82 were infected into PC3, DU145 and LNCaP. RT–qPCR values were compared to cells infected with a short hairpin control (shct) and results are presented as mean±s.e.m. of two biological replicates. (c) Representative pictures of the three cell lines infected either with shct or the two shs for CD82 in culture. (d) PC3, DU145 and LNCaP infected with either shct or the two shs for CD82 were plated in the presence of fibronectin or laminin I. An adhesion assay was again performed as previously described in Figure 4. Results are presented as mean±s.e.m. of three technical replicates of one representative biological replicate of two attempts.
Discussion
PC is one of the main causes of cancer death of men in western countries, with metastasis to the bone representing the predominant incurable last step in the pathogenesis of the tumor. Indeed, the metastatic process is a complex multistep process, including EMT and invasion from the primary tumor, dissemination through the blood and lymphatic vasculature, invasion into the metastatic organ, colonization and survival of the invading cells, culminating in the growth of the metastatic tumor. In the absence of suitable genetic models to investigate this process in PC, many studies rely on the PC3 cell line, which exhibits metastasis to the bone (and other tissues) in a subset of mice following intra-cardiac injection. Interestingly, this cell line also contains populations of cancer stem-like cells that favor clonal growth and tumor growth.11, 12, 13 Our results uncover a previously unknown sub-population of p63-positive cells in PC3, and identify a role for ΔNp63α in mediating prostate metastatic colonization to the bone and favoring the growth of the tumor in this microenvironment (Figure 7). Importantly, this seems to be through increased adhesion of tumor cells in the bone metastatic niche and the promotion of stem-like growth properties. The concept that metastatic colonization involves stem-like events has previously been demonstrated in breast cancer,34, 35, 36 and our study now suggests similar mechanisms also function in PC.
Model of p63 expression and function in PSCs and cancer. ΔNp63α+ basal PSCs give rise to ΔNp63α− luminal differentiated cells and can be targeted for transformation generating mainly luminal cells that form the bulk of the primary tumor. Loss of ΔNp63 is necessary to undergo EMT, enter the circulation and invade distant sites. Few cells survive these processes and in PC, they predominantly colonize the bone where they re-acquire an epithelial/stem cell phenotype through mesenchymal–epithelial transition and colonize the new microenvironment. Our data suggest that this step requires reactivation of ΔNp63α that drives the expression of molecules, such as CD82, necessary for adhesion to the bone. ΔNp63α- cells could then derive from ΔNp63α+ colonizing cells and participate to the formation of the metastatic lesions together with stromal and other bone resident cells.
ΔNp63α is a well-characterized mediator of stem cell function in a number of tissues, particularly during embryonic development.2, 37 We demonstrate that it is the utilization of some of the same regulatory pathways and processes mediated by ΔNp63α in normal stem cells that favors bone metastatic tumor growth. These processes include adhesion of the colonizing cell, signaling to the local microenvironment to create a niche and the likely recruitment of cooperating cell populations. Intriguingly, the fact that some of the p63-regulated pathways that help maintain normal PSC growth facilitate survival in the bone microenvironment might help to explain the predominance of metastasis to the bone with these tumors.
Moreover, this discovery that ΔNp63α is expressed in the bone metastatic cells and in the metastatic tumors, while being clinically absent from primary tumors, raises the possibility that the bone microenvironment might favor the re-expression of p63 at different stages of tumor progression. Interestingly, ΔNp63α-positive cells have been suggested as the cell of origin for PC,6, 7 while, loss of p63 expression is important for EMT and tumor invasion.7, 17, 18, 19, 27 In addition, its forced re-expression is sufficient to re-activate epithelial traits in a mesenchymal–epithelial transition-like process.19, 38 Our findings support these observations, that reactivation of ΔNp63α in a subset of metastatic cells at later stages of colonization might promote mesenchymal–epithelial transition and as we show, adhesion at bone metastatic sites (Figure 7). Indeed, our data adds to the growing body of work linking p63 with adhesion and tumor progression,27, 28, 29 and uncovers an unexpected role for this process in prostate bone metastatic tumor growth.
While the bone microenvironment may play a role in reactivation of p63, why there exists a small sub-population of p63-positive cells in this the PC3 cell line and not the others remains unknown. Here it may be related to the fact that these cells are androgen independent. In normal prostate, removal of androgen stimulation through castration causes regression of the prostate. However, this also causes an accumulation of p63-positive cells in the regressed gland, which are required for maintenance in the absence of hormone, and gland regrowth upon hormone restoration.39, 40 It is tempting to speculate that this enrichment of p63 in the absence of hormone may be related to the expression of p63 in the PC3 cell line. Of course, while the DU145 line also is androgen independent, PC3 retains the expression of coactivators necessary to mediate a response to hormone that is not present in the DU145 line,41 suggesting there are additional difference with regards to androgen independence between the two lines.
In addition, we also identified CD82, a known p63-target gene25 as being common to normal stem-like and metastatic prostate cells, and demonstrated that regulated interaction with the external environment through adhesion and adhesion-related signaling plays a significant role in regulating metastatic colonization.34, 35, 36, 42 Previously, CD82 has been well characterized as a metastasis suppressor at early stages of tumor development, a promoter of adhesion and a suppressor of EMT in a variety of primary tumors.30, 32, 43, 44 This is in part through the regulation of integrin-α4 stability,43 which was also enriched in ΔNp63α and CD82-high cells in the current study. However, our data suggest that CD82 can also act as a promoter of the later stages of metastasis, and specifically of metastatic colonization of the target tissue. Importantly, our findings suggest that targeted interruption of the protein might actually decrease metastatic burden. Interestingly, CD82 has previously been linked with hematopoietic and leukemic stem cell adhesion and homing.31, 45, 46 Our identification here of a sub-population of metastatic PC cells with stem-like properties and increased adhesion raises the interesting possibility that CD82 may also play a role in PC stem cell growth, as the adhesion properties conferred by CD82 were confirmed in the two other main cell line used to model PC metastasis, DU145 and LNCaP.
Altogether, the unexpected identification here of a sub-population of ΔNp63 and CD82-positive cells, whose disruption significantly perturbs the development of prostate metastatic tumor growth, further supports that targeting the interaction with the metastatic niche might represent a viable therapeutic opportunity at later stages of tumor progression.
Materials and methods
Animal studies
Male C57BL/6 and Swiss Nude mice of 6–10 weeks old (Charles Rivers Laboratories, Wilmington, MA, USA) were maintained at the Barcelona Biomedical Research Park, (PRBB), following the animal care standards of the institution that complies with European legislation on the care and use of animals. For intra-cardiac injections, 1 × 106 cells were resuspended in 100 μl of phosphate-buffered saline (PBS) and injected into the left ventricle of nude mice. Luciferase signal was acquired after 30 min and mice without a systemic distribution of the signal were discarded. For intra-tibial injections, 2.5 × 105 cells were resuspended in 20 μl of PBS and injected into the right tibias of nude mice. Left tibias were injected the same way with PBS as control. For imaging, 400 μl of 5 mg/ml potassium luciferin (GOLDBIO Cat#LUK1G) were injected intraperitoneally in each animal and signal was acquired after 10 min with the in vitro imaging system (IVIS) (Perkin Elmer) platform. Images were analyzed with the Igor Pro Carbon software. Luciferase signal is represented as mean±s.e.m. The tumor incidence refers to the total number of mice developing lesions from the beginning of the study, P-values are calculated using the log-rank test.
ShRNA design and cloning
For shRNA design, http://gesteland.genetics.utah.edu/siRNA_scales/ and http://cancan.cshl.edu/RNAi_central/RNAi.cgi?type=shRNA were used. The oligonucleotides were purchased from Invitrogen (Carlsbad, CA, USA) and were cloned in the pLMP-GFP-Puro retroviral expression vector.47 The sequence of the shΔNp63 used in our studies is 5′-TGCTGTTGACAGTGAGCGCAAGGACAGCAGCATTGATCAATAGTGAAGCCACAGATGTATTGATCAATGCTGCTGTCCTTTTGCCTACTGCCTCGGA-3′. For knockdown of CD82, five CD82 hairpins and a short hairpin control were purchased from Sigma-Aldrich (St Louis, MO, USA): TRCN0000245060, TRCN0000038029, TRCN0000038030, TRCN0000038032, TRCN0000038033 and SHC002 in the pLKO.1-puro vector. For overexpression studies, the retroviral control vector pMSCV-GFP-Puro and pMSCV-GFP-Puro containing full-length mouse ΔNp63α vectors were used.5 pMSCV-Luc-GFP-Neo was a gift from Johannes Zuber, IMP, Vienna.
Cell lines, viral production and infection
RWPE-1 cells were purchased from ATCC (Manassas, VA, USA). PC3, LNCaP and DU145 were a kind gift of Maria Paola Paronetto, Fondazione Santa Lucia, Universitá degli studi di Roma ‘Foro Italico’. Phoenix packaging cells (G Nolan, Stanford University, Stanford, CA, USA) were used for retrovirus production following previously described transfection procedures.5
Focus-formation assay, colony-formation assay and 3D culture
For focus-formation assay, 2000, 4000 and 8000 cells were cultured in duplicate in six-well plates. At day 10, cells were fixed and stained with 0,5% crystal violet in 20% MetOH. For colony-formation assay, 100 cells were cultured in triplicate in six-well plates and at 2 weeks, the cells were fixed and stained as before. P-values were calculated on three biological replicates using unpaired t-test for the number of colonies and two-way Anova with Bonferroni post-test for the percentage of holoclones, meroclones and paraclones. 3D cultures were performed as previously described.48, 49 2.5 × 103 cells were seeded into eight-well chamber slides (BD Falcon, Corning, NY, USA). Values from five and three experiments, respectively, for P1 and P2, are combined for quantification of number and area of at least 200 spheres/8 fields of view per condition per experiment.
The imaging software ImageJ was used for analysis. Statistical significance was determined by two-tailed Student’s t-test.
Adhesion assay
Forty-eight-well plates were coated with collagen | 1:100, fibronectin 20 μg/ml, laminin | 10 μg/ml (R&D systems, Minneapolis, MN, USA). Cells were plated at a concentration of 4 × 105/ml in a volume of 250 μl per well. At 30 min, 1 and 2 h after plating, adherent cells were fixed and attached cells were stained with 0,5% crystal violet in 20% MetOH for 10 min and counted with a × 40 objective in five randomly chosen fields per three wells for each condition. Statistics were performed using one-way Anova and Tukey’s multiple comparisons tests.
Flow cytometry
The antibodies for Sca-1 (FITC, E-13-161.7, BD Pharmigen, San Diego, CA, USA) and CD49f (PE, GoH3, BD Pharmigen) were used at a concentration of 1:100. CD82 was detected using the B-L2 antibody (Abcam, Cambridge, UK) and a secondary antibody conjugated with Alexa Fluor 488 or 568 (Invitrogen), respectively, 1:100 and 1:500. Cells were analyzed on the LSR Fortessa flow cytometer or sorted on the FACSAria cell sorter (BD Biosciences, San Diego, CA, USA) and data examined with FlowJo software (Tree Star, Ashland, OR, USA).
Cytospin
Prostates from 6–10-week-old wild-type C57BL/6 mice and were incubated in 5 mg/ml Dispase || (Roche, Basel, Switzerland) at 4 °C o/n. The tissue was minced and stirred for 30 min at RT. Dissociated cells were sorted and attached to a slide by spinning in a cytospin 4 centrifuge (Thermo Shandon, Cambridge, UK). For quantification after immunocytochemistry, cells were counted and statistics were performed using two-way Anova with Bonferroni post-test
Immunocytochemistry
Cells were washed, fixed and incubated 1 h at RT with blocking solution: PBS, 10% goat serum, 0.5% Tween 20 (Sigma-Aldrich). Primary and secondary antibodies were incubated, respectively, o/n at 4 °C and 1 h at RT. Primary antibody for p63 (4A4) was used at a concentration of 1:200. Secondary antibody conjugated with Alexa Fluor 568 (Invitrogen) was used 1:1000.
Histological sample processing, immunofluorescent and immunohistochemical stainings
Bones were fixed in 4% PFA o/n at 4 °C and decalcified in 10% EDTA at 4 °C for 3 weeks. All the samples were embedded in paraffin and sectioned. Antigen retrieval was performed by boiling the sections for 20 min in EDTA buffer and cooling to RT for 1 h.
For immunohistochemistry, sections were treated with 0.3% H2O2 in PBS for 15 min at RT. Permeabilization was performed with 0.2% Tween 20 (Sigma-Aldrich) for 5 min at RT and blocking solution (10% goat serum, 0.1% Tween 20 in PBS) was added for 1 h at RT. Primary and secondary antibodies were incubated, respectively, o/n at 4 °C and 1 h at RT. Antibodies for CK5 (H40, Santa Cruz Biotechnology, Dallas, TX, USA), CK18 (H-80, Santa Cruz Biotechnology) and ΔNp63 (p40 5–17, Millipore, Darmstadt, Germany) were used, respectively, at concentration 1:500, 1:100 and 1:1000.
For immunofluorescence, secondary antibodies conjugated with Alexa Fluor 488 or 568 (Invitrogen) were used 1:1000. For immunohictochemistry, biotin-coupled secondary antibody incubation and avidin-peroxidase label were performed with the ABC Kit (Vectastain, Vector Labs, Burlingame, CA, USA) and reactions were developed with DAB substrate kit for peroxidase (Vector Laboratories, Burlingame, CA, USA) according to the manufacturer’s instructions. Hematoxylin counterstain was used for nuclei.
RT–qPCR
Total RNA was isolated using TRIzol Reagent (Life Technologies) or the RNeasy Mini Kit (QIAGEN, Hilden, Germany). One microgram of RNA was reverse transcribed using the SuperScript ||| First-Strand kit from Invitrogen or the qScript cDNA SuperMix from Quanta, in a reaction volume of 20 μl (50 ng/μl), which was diluted 1:10 (5 ng/μl), with 2 μl of this dilution (10 ng) were used for qRT–PCR. qRT–PCR reactions were performed in triplicate using SYBR Green Master Mix and a Light Cycler 480 Instrument (Roche Life Science, Basel, Switzerland). All kits were used according to the manufacturers’ instructions. Relative levels of expression were determined relative to the housekeeping gene β-actin. Primers are listed in Supplementary Table 2. Three biological replicates were done for each experiment, unless differently specified. Results are presented in graphs as mean±s.e.m. Statistical significance was determined by one-sample t-test.
Western blot and IP
For IPs and western blots, cells were lysed in IP300 buffer, sonicated 15 s at 10% output using a Branson sonicator and centrifuged for 20 min at maximum speed. IPs for p63 were performed using 50 μl of Protein A Sepharose 4 Fast Flow (GE Healthcare, Little Chalfont, UK) and 5 μg of the antibody clone H-137 from Santa Cruz. Control IPs were performed similarly using normal mouse IgG sc-2025 (Santa Cruz Biotechnology). An aliquot of 0.5 mg of RWPE-1 lysate and 4 mg of PC3, LNCAP and DU145 protein lysates were used for IPs. Western blot membranes were stained for p63 (4A4) (Santa Cruz Biotechnology) at a concentration of 1:200 o/n at 4 °C. α-tubulin was used as loading control and detected with the DM1A antibody (Sigma-Aldrich) at a concentration of 1:5000 1 h at RT. Anti-mouse IgG (GE Healthcare) was used as secondary antibody at a concentration of 1:5000 1 h at RT.
Microarray analysis and ChIP
For microarray analysis on RWPE-1 cells with knockdown of ΔNp63, three and four technical replicates of R-V and R-shΔN group were analyzed, respectively. For PC3 cells, two technical replicates were analyzed for P-V and four for P-ΔNα. Total RNA was extracted with the RNeasy Mini Kit (QIAGEN) and 100 ng were labeled using Low Input Quick Amp Labeling kit v6.5 (Agilent Technologies, Santa Clara, CA, USA; 5190-2305) following the manufacturer’s instructions.
Chromatin IP was performed on RWPE-1 cells adapting the protocol from Keyes et al.5 The 4A4 antibody for p63 (Santa Cruz, SC-8431) was used. Libraries were prepared using the NEBNext ChIP-Seq Library Prep Reagent Set for Illumina kit (ref. E6200S) according to the manufacturer’s protocols.
References
Yang A, Kaghad M, Wang Y, Gillett E, Fleming MD, Dotsch V et al. p63, a p53 homolog at 3q27-29, encodes multiple products with transactivating, death-inducing, and dominant-negative activities. Mol Cell 1998; 2: 305–316.
Crum CP, McKeon FD . p63 in epithelial survival, germ cell surveillance, and neoplasia. Annu Rev Pathol 2010; 5: 349–371.
Graziano V, De Laurenzi V . Role of p63 in cancer development. Biochim Biophys Acta 2011; 1816: 57–66.
Hibi K, Trink B, Patturajan M, Westra WH, Caballero OL, Hill DE et al. AIS is an oncogene amplified in squamous cell carcinoma. Proc Natl Acad Sci USA 2000; 97: 5462–5467.
Keyes WM, Pecoraro M, Aranda V, Vernersson-Lindahl E, Li W, Vogel H et al. ΔNp63α is an oncogene that targets chromatin remodeler Lsh to drive skin stem cell proliferation and tumorigenesis. Cell Stem Cell 2011; 8: 164–176.
Goldstein AS, Huang J, Guo C, Garraway IP, Witte ON . Identification of a cell of origin for human prostate cancer. Science 2010; 329: 568–571.
Wang S, Garcia AJ, Wu M, Lawson DA, Witte ON, Wu H . Pten deletion leads to the expansion of a prostatic stem/progenitor cell subpopulation and tumor initiation. Proc Natl Acad Sci USA 2006; 103: 1480–1485.
Di Como CJ, Urist MJ, Babayan I, Drobnjak M, Hedvat CV, Teruya-Feldstein J et al. p63 expression profiles in human normal and tumor tissues. Clin Cancer Res 2002; 8: 494–501.
Grisanzio C, Signoretti S . p63 in prostate biology and pathology. J Cell Biochem 2008; 103: 1354–1368.
Signoretti S, Waltregny D, Dilks J, Isaac B, Lin D, Garraway L et al. p63 is a prostate basal cell marker and is required for prostate development. Am J Pathol 2000; 157: 1769–1775.
Doherty RE, Haywood-Small SL, Sisley K, Cross NA . Aldehyde dehydrogenase activity selects for the holoclone phenotype in prostate cancer cells. Biochem Biophys Res Commun 2011; 414: 801–807.
Li H, Chen X, Calhoun-Davis T, Claypool K, Tang DG . PC3 human prostate carcinoma cell holoclones contain self-renewing tumor-initiating cells. Cancer Res 2008; 68: 1820–1825.
Zhang K, Waxman DJ . PC3 prostate tumor-initiating cells with molecular profile FAM65Bhigh/MFI2low/LEF1low increase tumor angiogenesis. Mol Cancer 2010; 9: 319.
Mills AA, Zheng B, Wang XJ, Vogel H, Roop DR, Bradley A . p63 is a p53 homologue required for limb and epidermal morphogenesis. Nature 1999; 398: 708–713.
Signoretti S, Pires MM, Lindauer M, Horner JW, Grisanzio C, Dhar S et al. p63 regulates commitment to the prostate cell lineage. Proc Natl Acad Sci USA 2005; 102: 11355–11360.
Yang A, Schweitzer R, Sun D, Kaghad M, Walker N, Bronson RT et al. p63 is essential for regenerative proliferation in limb, craniofacial and epithelial development. Nature 1999; 398: 714–718.
Mulholland DJ, Xin L, Morim A, Lawson D, Witte O, Wu H . Lin-Sca-1+CD49fhigh stem/progenitors are tumor-initiating cells in the Pten-null prostate cancer model. Cancer Res 2009; 69: 8555–8562.
Lawson DA, Zong Y, Memarzadeh S, Xin L, Huang J, Witte ON . Basal epithelial stem cells are efficient targets for prostate cancer initiation. Proc Natl Acad Sci USA 2010; 107: 2610–2615.
Tucci P, Agostini M, Grespi F, Markert EK, Terrinoni A, Vousden KH et al. Loss of p63 and its microRNA-205 target results in enhanced cell migration and metastasis in prostate cancer. Proc Natl Acad Sci USA 2012; 109: 15312–15317.
Chae YS, Kim H, Kim D, Lee H, Lee HO . Cell density-dependent acetylation of ΔNp63α is associated with p53-dependent cell cycle arrest. FEBS Lett 2012; 586: 1128–1134.
Park SI, Kim SJ, McCauley LK, Gallick GE . Pre-clinical mouse models of human prostate cancer and their utility in drug discovery. Curr Protoc Pharmacol 2010; Chapter 14: Unit 14. 15.
Valkenburg KC, Williams BO . Mouse models of prostate cancer. Prostate Cancer 2011; 2011: 895238.
Liu Q, Russell MR, Shahriari K, Jernigan DL, Lioni MI, Garcia FU et al. Interleukin-1beta promotes skeletal colonization and progression of metastatic prostate cancer cells with neuroendocrine features. Cancer Res 2013; 73: 3297–3305.
Osada M, Nagakawa Y, Park HL, Yamashita K, Wu G, Kim MS et al. p63-specific activation of the BPAG-1e promoter. J Invest Dermatol 2005; 125: 52–60.
Wu J, Liang S, Bergholz J, He H, Walsh EM, Zhang Y et al. ΔNp63α activates CD82 metastasis suppressor to inhibit cancer cell invasion. Cell Death Dis 2014; 5: e1280.
Wu N, Rollin J, Masse I, Lamartine J, Gidrol X . p63 regulates human keratinocyte proliferation via MYC-regulated gene network and differentiation commitment through cell adhesion-related gene network. J Biol Chem 2012; 287: 5627–5638.
Barbieri CE, Tang LJ, Brown KA, Pietenpol JA . Loss of p63 leads to increased cell migration and up-regulation of genes involved in invasion and metastasis. Cancer Res 2006; 66: 7589–7597.
Carroll DK, Carroll JS, Leong CO, Cheng F, Brown M, Mills AA et al. p63 regulates an adhesion programme and cell survival in epithelial cells. Nat Cell Biol 2006; 8: 551–561.
Ferone G, Mollo MR, Missero C . Epidermal cell junctions and their regulation by p63 in health and disease. Cell Tissue Res 2015; 360: 513–528.
Dong JT, Lamb PW, Rinker-Schaeffer CW, Vukanovic J, Ichikawa T, Isaacs JT et al. KAI1, a metastasis suppressor gene for prostate cancer on human chromosome 11p11.2. Science 1995; 268: 884–886.
Nishioka C, Ikezoe T, Furihata M, Yang J, Serada S, Naka T et al. CD34(+)/CD38(−) acute myelogenous leukemia cells aberrantly express CD82 which regulates adhesion and survival of leukemia stem cells. Int J Cancer 2013; 132: 2006–2019.
Tsai YC, Weissman AM . Dissecting the diverse functions of the metastasis suppressor CD82/KAI1. FEBS Lett 2011; 585: 3166–3173.
Lee HA, Park I, Byun HJ, Jeoung D, Kim YM, Lee H . Metastasis suppressor KAI1/CD82 attenuates the matrix adhesion of human prostate cancer cells by suppressing fibronectin expression and beta1 integrin activation. Cell Physiol Biochem 2011; 27: 575–586.
Gupta GP, Perk J, Acharyya S, de Candia P, Mittal V, Todorova-Manova K et al. ID genes mediate tumor reinitiation during breast cancer lung metastasis. Proc Natl Acad Sci USA 2007; 104: 19506–19511.
Malanchi I, Santamaria-Martinez A, Susanto E, Peng H, Lehr HA, Delaloye JF et al. Interactions between cancer stem cells and their niche govern metastatic colonization. Nature 2012; 481: 85–89.
Oskarsson T, Acharyya S, Zhang XH, Vanharanta S, Tavazoie SF, Morris PG et al. Breast cancer cells produce tenascin C as a metastatic niche component to colonize the lungs. Nat Med 2011; 17: 867–874.
Senoo M, Pinto F, Crum CP, McKeon F . p63 Is essential for the proliferative potential of stem cells in stratified epithelia. Cell 2007; 129: 523–536.
Olsen JR, Oyan AM, Rostad K, Hellem MR, Liu J, Li L et al. p63 attenuates epithelial to mesenchymal potential in an experimental prostate cell model. PLoS One 2013; 8: e62547.
Kurita T, Medina RT, Mills AA, Cunha GR . Role of p63 and basal cells in the prostate. Development 2004; 131: 4955–4964.
Tien JC, Liu Z, Liao L, Wang F, Xu Y, Wu YL et al. The steroid receptor coactivator-3 is required for the development of castration-resistant prostate cancer. Cancer Res 2013; 73: 3997–4008.
Litvinov IV, Antony L, Dalrymple SL, Becker R, Cheng L, Isaacs JT . PC3, but not DU145, human prostate cancer cells retain the coregulators required for tumor suppressor ability of androgen receptor. Prostate 2006; 66: 1329–1338.
Seguin L, Desgrosellier JS, Weis SM, Cheresh DA . Integrins and cancer: regulators of cancer stemness, metastasis, and drug resistance. Trends Cell Biol 2015; 25: 234–240.
Termini CM, Cotter ML, Marjon KD, Buranda T, Lidke KA, Gillette JM . The membrane scaffold CD82 regulates cell adhesion by altering α4 integrin stability and molecular density. Mol Biol Cell 2014; 25: 1560–1573.
Tonoli H, Barrett JC . CD82 metastasis suppressor gene: a potential target for new therapeutics? Trends Mol Med 2005; 11: 563–570.
Burchert A, Notter M, Dietrich Menssen H, Schwartz S, Knauf W, Neubauer A et al. CD82 (KAI1), a member of the tetraspan family, is expressed on early haemopoietic progenitor cells and up-regulated in distinct human leukaemias. Br J Haematol 1999; 107: 494–504.
Larochelle A, Gillette JM, Desmond R, Ichwan B, Cantilena A, Cerf A et al. Bone marrow homing and engraftment of human hematopoietic stem and progenitor cells is mediated by a polarized membrane domain. Blood 2012; 119: 1848–1855.
Dickins RA, Hemann MT, Zilfou JT, Simpson DR, Ibarra I, Hannon GJ et al. Probing tumor phenotypes using stable and regulated synthetic microRNA precursors. Nat Genet 2005; 37: 1289–1295.
Aranda V, Haire T, Nolan ME, Calarco JP, Rosenberg AZ, Fawcett JP et al. Par6-aPKC uncouples ErbB2 induced disruption of polarized epithelial organization from proliferation control. Nat Cell Biol 2006; 8: 1235–1245.
Harma V, Virtanen J, Makela R, Happonen A, Mpindi JP, Knuuttila M et al. A comprehensive panel of three-dimensional models for studies of prostate cancer growth, invasion and drug responses. PLoS One 2010; 5: e10431.
Acknowledgements
We thank Roger Gomis and Marc Guiu for help with tumor imaging analysis, Jason Doles for help in the short hairpin design and the CRG Bioinformatics facility for assistance with microarray and sequencing data analysis. VDG was supported by a fellowship from ‘La Caixa’, and MP with an F.P.I. fellowship from the Spanish Ministry. This work was funded by Grants SAF2010-18829 (WMK) and SAF2013-49082-P (WMK) from the Ministerio de Economía y Competitividad and a Marie Curie International Reintegration Grant PIRG5-GA-2009-248262 (WMK).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Oncogene website
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/4.0/
About this article
Cite this article
Di Giacomo, V., Tian, T., Mas, A. et al. ΔNp63α promotes adhesion of metastatic prostate cancer cells to the bone through regulation of CD82. Oncogene 36, 4381–4392 (2017). https://doi.org/10.1038/onc.2017.42
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/onc.2017.42
This article is cited by
-
Tumor- and osteoclast-derived NRP2 in prostate cancer bone metastases
Bone Research (2021)
-
TAp63 and ΔNp63 (p40) in prostate adenocarcinomas: ΔNp63 associates with a basal-like cancer stem cell population but not with metastasis
Virchows Archiv (2021)
-
TP63 links chromatin remodeling and enhancer reprogramming to epidermal differentiation and squamous cell carcinoma development
Cellular and Molecular Life Sciences (2020)
-
Expression of tetraspanins NET-6 and CD151 in breast cancer as a potential tumor biomarker
Clinical and Experimental Medicine (2019)