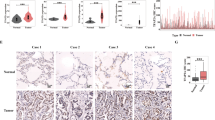
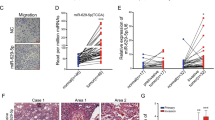
Abstract
MicroRNAs (miRNAs) have been proposed as critical regulatory molecules in the epithelial–mesenchymal transition (EMT) program. However, the roles of mature miRNA biogenesis during EMT process needs to be defined. Here we determined that increased expression of XRN2 induced EMT and promoted metastasis in vitro and in vivo. Furthermore, we uncovered that XRN2 functions as pro-metastatic gene, which accelerates miR-10a maturation by binding pre-miR-10a in a DICER-independent manner. These findings suggest that XRN2 is a novel regulator of EMT that contributes to the metastatic processes in lung cancer through a novel miRNA regulatory mechanism.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Hanahan D, Weinberg RA . Hallmarks of cancer: the next generation. Cell 2011; 144: 646–674.
Tsai JH, Yang J . Epithelial-mesenchymal plasticity in carcinoma metastasis. Genes Dev 2013; 27: 2192–2206.
Tam WL, Weinberg RA . The epigenetics of epithelial-mesenchymal plasticity in cancer. Nat Med 2013; 19: 1438–1449.
Nieto MA . Epithelial plasticity: a common theme in embryonic and cancer cells. Science 2013; 342: 1234850.
Acloque H, Adams MS, Fishwick K, Bronner-Fraser M, Nieto MA . Epithelial-mesenchymal transitions: the importance of changing cell state in development and disease. J Clin Invest 2009; 119: 1438–1449.
Thiery JP, Acloque H, Huang RY, Nieto MA . Epithelial-mesenchymal transitions in development and disease. Cell 2009; 139: 871–890.
Zhang J, Ma L . MicroRNA control of epithelial-mesenchymal transition and metastasis. Cancer Metastasis Rev 2012; 31: 653–662.
Zhang H, Li Y, Lai M . The microRNA network and tumor metastasis. Oncogene 2010; 29: 937–948.
Gregory PA, Bert AG, Paterson EL, Barry SC, Tsykin A, Farshid G et al. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol 2008; 10: 593–601.
Park SM, Gaur AB, Lengyel E, Peter ME . The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev 2008; 22: 894–907.
Korpal M, Lee ES, Hu G, Kang Y . The miR-200 family inhibits epithelial-mesenchymal transition and cancer cell migration by direct targeting of E-cadherin transcriptional repressors ZEB1 and ZEB2. J Biol Chem 2008; 283: 14910–14914.
Ebert MS, Sharp PA . Roles for microRNAs in conferring robustness to biological processes. Cell 2012; 149: 515–524.
Miki TS, Grosshans H . The multifunctional RNase XRN2. Biochem Soc Trans 2013; 41: 825–830.
Wang M, Pestov DG . 5'-end surveillance by Xrn2 acts as a shared mechanism for mammalian pre-rRNA maturation and decay. Nucleic Acids Res 2011; 39: 1811–1822.
Zakrzewska-Placzek M, Souret FF, Sobczyk GJ, Green PJ, Kufel J . Arabidopsis thaliana XRN2 is required for primary cleavage in the pre-ribosomal RNA. Nucleic Acids Res 2010; 38: 4487–4502.
Couvillion MT, Bounova G, Purdom E, Speed TP, Collins K . A Tetrahymena Piwi bound to mature tRNA 3' fragments activates the exonuclease Xrn2 for RNA processing in the nucleus. Mol Cell 2012; 48: 509–520.
Lu Y, Liu P, James M, Vikis HG, Liu H, Wen W et al. Genetic variants cis-regulating Xrn2 expression contribute to the risk of spontaneous lung tumor. Oncogene 2010; 29: 1041–1049.
Bild AH, Yao G, Chang JT, Wang Q, Potti A, Chasse D et al. Oncogenic pathway signatures in human cancers as a guide to targeted therapies. Nature 2006; 439: 353–357.
Kuner R, Muley T, Meister M, Ruschhaupt M, Buness A, Xu EC et al. Global gene expression analysis reveals specific patterns of cell junctions in non-small cell lung cancer subtypes. Lung Cancer 2009; 63: 32–38.
Tomida S, Takeuchi T, Shimada Y, Arima C, Matsuo K, Mitsudomi T et al. Relapse-related molecular signature in lung adenocarcinomas identifies patients with dismal prognosis. J Clin Oncol 2009; 27: 2793–2799.
Lu Y, Lemon W, Liu PY, Yi Y, Morrison C, Yang P et al. A gene expression signature predicts survival of patients with stage I non-small cell lung cancer. PLoS Med 2006; 3: e467.
Chatterjee S, Grosshans H . Active turnover modulates mature microRNA activity in Caenorhabditis elegans. Nature 2009; 461: 546–549.
Lu Y, Govindan R, Wang L, Liu PY, Goodgame B, Wen W et al. MicroRNA profiling and prediction of recurrence/relapse-free survival in stage I lung cancer. Carcinogenesis 2012; 33: 1046–1054.
Xiao F, Zuo Z, Cai G, Kang S, Gao X, Li T . miRecords: an integrated resource for microRNA-target interactions. Nucleic Acids Res 2009; 37: D105–D110.
Zhao M, Kong L, Liu Y, Qu H . dbEMT: an epithelial-mesenchymal transition associated gene resource. Sci Rep 2015; 5: 11459.
Ma L, Teruya-Feldstein J, Weinberg RA . Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature 2007; 449: 682–688.
Shah N, Sukumar S . The Hox genes and their roles in oncogenesis. Nat Rev Cancer 2010; 10: 361–371.
Weiss FU, Marques IJ, Woltering JM, Vlecken DH, Aghdassi A, Partecke LI et al. Retinoic acid receptor antagonists inhibit miR-10a expression and block metastatic behavior of pancreatic cancer. Gastroenterology 2009; 137: 2136–2145 e2131-2137.
Chang CH, Fan TC, Yu JC, Liao GS, Lin YC, Shih A et al. The prognostic significance of RUNX2 and miR-10a/10b and their inter-relationship in breast cancer. J Transl Med 2014; 12: 257.
Ohuchida K, Mizumoto K, Lin C, Yamaguchi H, Ohtsuka T, Sato N et al. MicroRNA-10a is overexpressed in human pancreatic cancer and involved in its invasiveness partially via suppression of the HOXA1 gene. Ann Surg Oncol 2012; 19: 2394–2402.
Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci USA 2006; 103: 2257–2261.
Varnholt H, Drebber U, Schulze F, Wedemeyer I, Schirmacher P, Dienes HP et al. MicroRNA gene expression profile of hepatitis C virus-associated hepatocellular carcinoma. Hepatology 2008; 47: 1223–1232.
Dip N, Reis ST, Timoszczuk LS, Viana NI, Piantino CB, Morais DR et al. Stage, grade and behavior of bladder urothelial carcinoma defined by the microRNA expression profile. J Urol 2012; 188: 1951–1956.
Lund AH . miR-10 in development and cancer. Cell Death Differ 2010; 17: 209–214.
Long MJ, Wu FX, Li P, Liu M, Li X, Tang H . MicroRNA-10a targets CHL1 and promotes cell growth, migration and invasion in human cervical cancer cells. Cancer Lett 2012; 324: 186–196.
Li X, Xu F, Chang C, Byon J, Papayannopoulou T, Deeg HJ et al. Transcriptional regulation of miR-10a/b by TWIST-1 in myelodysplastic syndromes. Haematologica 2013; 98: 414–419.
Yan Y, Luo YC, Wan HY, Wang J, Zhang PP, Liu M et al. MicroRNA-10a is involved in the metastatic process by regulating Eph tyrosine kinase receptor A4-mediated epithelial-mesenchymal transition and adhesion in hepatoma cells. Hepatology 2013; 57: 667–677.
Takahashi H, Kanno T, Nakayamada S, Hirahara K, Sciume G, Muljo SA et al. TGF-beta and retinoic acid induce the microRNA miR-10a, which targets Bcl-6 and constrains the plasticity of helper T cells. Nat Immunol 2012; 13: 587–595.
Fang Y, Shi C, Manduchi E, Civelek M, Davies PF . MicroRNA-10a regulation of proinflammatory phenotype in athero-susceptible endothelium in vivo and in vitro. Proc Natl Acad Sci USA 2010; 107: 13450–13455.
Miki TS, Ruegger S, Gaidatzis D, Stadler MB, Grosshans H . Engineering of a conditional allele reveals multiple roles of XRN2 in Caenorhabditis elegans development and substrate specificity in microRNA turnover. Nucleic Acids Res 2014; 42: 4056–4067.
Ha M, Kim VN . Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol 2014; 15: 509–524.
Xiang S, Cooper-Morgan A, Jiao X, Kiledjian M, Manley JL, Tong L . Structure and function of the 5'→3' exoribonuclease Rat1 and its activating partner Rai1. Nature 2009; 458: 784–788.
Wagschal A, Rousset E, Basavarajaiah P, Contreras X, Harwig A, Laurent-Chabalier S et al. Microprocessor, Setx, Xrn2, and Rrp6 co-operate to induce premature termination of transcription by RNAPII. Cell 2012; 150: 1147–1157.
Langmead B, Trapnell C, Pop M, Salzberg SL . Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 2009; 10: R25.
Robinson MD, McCarthy DJ, Smyth GK . edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010; 26: 139–140.
Benjamini Y, Hochberg Y . Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B (Methodological) 1995; 57: 289–300.
Li H, Gui J . Partial Cox regression analysis for high-dimensional microarray gene expression data. Bioinformatics 2004; 20 (Suppl 1): i208–i215.
Acknowledgements
This work has been support by National Natural Science Foundation of China (no. 81372514, 81472420, 81572256 and 31401125), National Key Research and Development program (2016YFA0501800 and 2016YFC0902700), the Fundamental Research Funds for the Central Universities and 1000 talent Plan of China, Louisiana Hope Research Grant provided by Free to Breathe. We thank Alison Kriegel for reading and commenting on the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Oncogene website
Supplementary information
Rights and permissions
About this article
Cite this article
Zhang, H., Lu, Y., Chen, E. et al. XRN2 promotes EMT and metastasis through regulating maturation of miR-10a. Oncogene 36, 3925–3933 (2017). https://doi.org/10.1038/onc.2017.39
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/onc.2017.39
This article is cited by
-
Midkine promotes glioblastoma progression via PI3K-Akt signaling
Cancer Cell International (2021)
-
High-throughput screening identified miR-7-2-3p and miR-29c-3p as metastasis suppressors in gallbladder carcinoma
Journal of Gastroenterology (2020)
-
Regulation of microRNA biogenesis and its crosstalk with other cellular pathways
Nature Reviews Molecular Cell Biology (2019)