Abstract
During normal tumor growth and in response to some therapies, tumor cells experience acute or chronic deprivation of nutrients and oxygen and induce tumor vascularization. While this occurs predominately through sprouting angiogenesis, tumor cells have also been shown to directly contribute to vessel formation through vascular mimicry (VM) and/or endothelial transdifferentiation. The extrinsic and intrinsic mechanisms underlying tumor cell adoption of endothelial phenotypes, however, are not well understood. Here we show that serum withdrawal induces mesenchymal breast cancer cells to undergo VM and that knockdown of the epithelial-to-mesenchymal transition (EMT) regulator, Zinc finger E-box binding homeobox 1 (ZEB1), or overexpression of the ZEB1-repressed microRNAs (miRNAs), miR-200c, miR-183, miR-96 and miR-182 inhibits this process. We find that secreted proteins Fibronectin 1 (FN1) and serine protease inhibitor (serpin) family E member 2 (SERPINE2) are essential for VM in this system. These secreted factors are upregulated in mesenchymal cells in response to serum withdrawal, and overexpression of VM-inhibiting miRNAs abrogates this upregulation. Intriguingly, the receptors for these secreted proteins, low-density lipoprotein receptor-related protein 1 (LRP1) and Integrin beta 1 (ITGB1), are also targets of the VM-inhibiting miRNAs, suggesting that autocrine signaling stimulating VM is regulated by ZEB1-repressed miRNA clusters. Together, these data provide mechanistic insight into the regulation of VM and suggest that miRNAs repressed during EMT, in addition to suppressing migratory and stem-like properties of tumor cells, also inhibit endothelial phenotypes of breast cancer cells adopted in response to a nutrient-deficient microenvironment.
Similar content being viewed by others
Introduction
As solid tumors increase in size, they deplete the local microenvironment of nutrients and oxygen. Tumor cells canonically respond to this stress by eliciting the recruitment of vasculature via sprouting angiogenesis.1 While the nascent tumor vessels are often disorganized and dysfunctional, they nevertheless resupply the oxygen and nutrients necessary to facilitate tumor growth, as well as provide a route by which tumor cells can metastasize.2 Inhibitors of angiogenesis have been developed for the treatment of tumors, but these have shown only modest effects on survival in many tumor types, including breast cancer.3 This has been linked, at least in part, to effects of the nutrient- and oxygen-deficient microenvironment resulting from these treatments on tumor cell phenotypes. For example, increased hypoxia following anti-angiogenic therapy can drive an epithelial-to-mesenchymal transition, which induces migratory and stem-like phenotypes in cancer cells.4, 5, 6 More recently, treatment with anti-angiogenic therapies has also been shown to increase alternative modes of vascularization, including tumor cell adoption of endothelial phenotypes through vascular mimicry (VM) and/or endothelial transdifferentiation.7, 8 While such alternative methods of tumor vascularization have been shown to impact tumor growth and metastasis,7, 9, 10 the extrinsic signals that drive and the intrinsic pathways that regulate these processes are poorly understood.
Here we utilize a manipulable, in vitro model of VM to show that mesenchymal breast cancer cells form endothelial-like networks when plated on Matrigel (BD Biosciences, San Jose, CA, USA) in the absence of serum. This VM is accompanied by enrichment, under network-forming conditions, of gene signatures expressed by endothelial cells in response to vascular endothelial growth factor or hypoxia. We find that knockdown of ZEB1 or re-expression of ZEB1-repressed microRNA (miRNA) clusters, miR-200c.141 or miR-183.96.182, is sufficient to inhibit VM and further, that transient expression of miR-200c, -183, -96 and -182, but not miR-141, blocks VM. We identify FN1 and the COPII secretory pathway machinery protein SEC23A as targets of miR-200c but not miR-141 in this system, suggesting a role for the cancer cell secretome in VM. We show that blocking COPII-mediated secretion through knockdown of SEC13 is sufficient to inhibit VM and, finally, we uncover a pathway whereby mesenchymal breast cancer cells, in response to serum withdrawal, upregulate secreted proteins FN1 and SERPINE2 (also called protease nexin 1) that are critical for VM. Specific miRNAs repressed during epithelial-to-mesenchymal transition can inhibit these secreted proteins as well as their cellular receptors ITGB1 and LRP1. High expression of the autocrine signaling factors involved in VM: FN1, ITGB1, SERPINE2, and LRP1, is found in claudin-low cancer cell lines and is significantly correlated with decreased survival in breast cancer patients. Together our data indicate that in addition to regulating migratory and stem-like properties of tumor cells, ZEB1-repressed miRNAs can also inhibit autocrine signaling that contributes to the ability of cancer cells to undergo VM in response to a nutrient-deficient microenvironment.
Results
Serum deprivation induces VM in mesenchymal breast cancer cell lines
VM has been associated with therapeutic resistance and metastasis in breast cancer.10, 11 To examine the intrinsic and extrinsic requirements for breast cancer cells to undergo VM, we utilized an in vitro VM assay that assesses cell network formation on Matrigel.12 As triple-negative (ER−PR−HER2−) breast cancer (TNBC) has been associated with VM,13, 14 we began by testing conditions under which the TNBC cell line, MDA-MB-231, would undergo VM. While we did not observe VM when MDA-MB-231 breast cancer cells were plated on top of Matrigel in normal growth media (10% serum), when these cells were plated in the absence of serum, the cells spread out across the Matrigel in a network resembling that seen with endothelial cord formation (Figure 1a). To determine whether this phenotype was characteristic of multiple TNBC cell lines, we additionally tested BT-549, Hs578T, SUM159PT, HCC1395, MDA-MB-453 and MDA-MB-468 cells in the serum-free VM assay. We found that four out of seven of the breast cancer cell lines, MDA-MB-231, BT-549, Hs578T and SUM159PT, were able to undergo network formation, while HCC1395, MDA-MB-453 and MDA-MB-468 cells were not (Figures 1b and c). The lines that we found to undergo VM belong to the mesenchymal-like or claudin-low subtype within TNBC.15, 16 We confirmed that the VM-competent lines express higher levels of epithelial-to-mesenchymal transition transcription factors, including ZEB1, and that only the MDA-MB-453 and MDA-MB-468 cell lines express E-cadherin (CDH1; Supplementary Figure S1A). In addition, flow cytometry analysis showed that the VM-competent lines MDA-MB-231 and BT-549 had substantially higher percentages of cells with CD44hi/CD24low expression as compared to MDA-MB-453 and MDA-MB-468 cell lines, suggesting the presence of a stem-like population associated with VM (Supplementary Figure S1B).
Serum deprivation induces VM of mesenchymal breast cancer cell lines. (a) Phase images of cells. MDA-MB-231 cells plated on (i) tissue culture plates in 10% serum, (ii) Matrigel in 10% serum or (iii) Matrigel in 0% serum, and (iv) HUVECs plated on Matrigel in EndoGRO (2% serum). Scale bars=400 μm. (b) Phase images of cells plated on Matrigel in 0% serum (VM assay). Scale bars=1000 μm. (c) Quantification of total network length was performed using the neurite tracing function within the NeuronGrowth plug-in on ImageJ59 over at least three independent replicates for each cell line. Error bars represent s.d. (d) Images of co-culture VM assay with HUVECs (red) and breast cancer cells (green). Phase images (top), fluorescent images (middle) and insets (bottom). Arrowheads indicate mosaic vessels and arrows indicate cancer cells not incorporated into networks. (e) Table of normalized enrichment scores (NES) and false discovery rate (FDR)-adjusted q-values from Gene Signature Enrichment Analysis for MDA-MB-231 cells plated on Matrigel in 0% vs 10% serum as compared to indicated gene signatures. A description of each of these gene signatures is listed in Supplementary Figure S1J.
The ability of the mesenchymal cell lines to form networks on Matrigel was inhibited by serum, as evidenced by a decrease in total network length exhibited by MDA-MB-231 and BT-549 cells plated in increasing amounts of serum (Supplementary Figures S1C–F). As tumor cells have been shown to form mosaic vessels with normal vasculature,17 we next performed co-culture assays with four of the breast cancer cell lines labeled with Cell Tracker Green and human umbilical vein endothelial cells (HUVECs) labeled with Cell Tracker Orange (Life Technologies, Carlsbad, CA, USA). These cells were plated together on Matrigel in EndoGRO media (Millipore, Temecula, CA, USA) containing 2% serum, a level that allows for cancer cell network formation (as shown in Supplementary Figures S1C–F), and networks were assessed 24 h later. VM-competent cell lines MDA-MB-231 and BT-549 formed mosaic cords with HUVECs on Matrigel, while the VM-incompetent lines MDA-MB-453 and MDA-MB-468 did not contribute to cord formation and remained distinct from the endothelial network (Figure 1d).
To gain mechanistic understanding of how breast cancer cells respond to growth factor depletion to form endothelial-like networks, we performed RNA-seq on MDA-MB-231 cells plated on Matrigel for 24 h in 10 or 0% serum. These data are publicly available in the National Center for Biotechnology Information Gene Expression Omnibus database (accession # GSE77179). We compared gene expression in 0% (network-forming) vs 10% (non-network-forming) serum (Supplementary Table 1) and used Gene Set Enrichment Analysis18 to determine gene programs that changed under network-forming conditions. Under the network-forming conditions, we found a significant enrichment in the expression of genes that are upregulated, and a de-enrichment in genes that are downregulated, in HUVECs upon treatment with vascular endothelial growth factor19 indicating mimicry of an angiogenic response (Figure 1e; Supplementary Figure S1G). In addition, we observed enrichment of upregulated and de-enrichment of downregulated signatures related to the response to hypoxia in arterial endothelial cells as well as other cell types (Figure 1e; Supplementary Figure S1H),20, 21, 22, 23 again suggesting mimicry of angiogenic pathways. Finally, we used Gene Set Enrichment Analysis to compare our RNA-seq data to the genes found in our previous work to be induced in embryonic stem cells that are differentiating into endothelial cells following induction of Snail (Flk1hi subpopulation).24 The MDA-MB-231 cells that undergo VM showed a significant enrichment of the Flk1hi differentiating endothelial cell signature (Figure 1e; Supplementary Figure S1I). Together, these data show that growth factor deprivation can induce VM in mesenchymal breast cancer cells and that breast cancer cells that undergo VM upregulate both angiogenic and endothelial differentiation pathways during this process.
ZEB1-regulated miRNA clusters block VM
Because the VM phenotype in response to growth factor deprivation appeared specific to mesenchymal cells, we first tested whether loss of the mesenchymal phenotype would inhibit VM. For this, we chose to knockdown ZEB1 as, unlike SNAI1, it was highly expressed in all of the VM-competent lines (Supplementary Figure S1A). We transfected MDA-MB-231 cells with pooled small interfering RNAs (siRNAs) against ZEB1 or a non-targeting control and confirmed knockdown at the mRNA and protein levels (Figures 2a and b). We then utilized these cells in the VM assay and found that knockdown of ZEB1 significantly inhibited network formation (Figures 2c and d). In our previous study of Snail-driven endothelial differentiation of embryonic stem cells, we identified miRNA clusters downregulated in this process, including miR-200c.141 and miR-183.96.18225 that both not only target but are also stably downregulated, by ZEB1.26, 27, 28, 29 These miRNA clusters have also been shown to be among the most repressed miRNAs in breast cancer stem cells.30, 31 Therefore, we sought to determine whether repression of these miRNA clusters by ZEB1 had a functional role in VM. First, we confirmed regulation of these miRNA clusters by ZEB1 and found that these miRNAs were indeed increased following a decrease in ZEB1 expression (Figure 2e). Next, we overexpressed miRNAs of each cluster in the MDA-MB-231 cell line, and confirmed expression by quantitative reverse transcription–PCR (Supplementary Figures S2A and B). Both combinations of miR-200c+miR-141 and miR-183+miR-96+miR-182 were able to completely block VM when overexpressed in mesenchymal breast cancer cells (Figures 2f and g). As expected, expression of miR-200c+miR-141 drove a mesenchymal to epithelial transition, evidenced by morphological changes, loss of ZEB1 and FN1 expression, and upregulation of CDH1 (Figures 2h–k).27, 28, 29 Expression of miR-183+miR-96+miR-182 also significantly decreased expression of ZEB1 and FN1, and upregulated CDH1, albeit to a lesser extent than miR-200c+miR-141 (Figures 2i–k). SNAI2 and VIM downregulation was also observed following expression of miR-183+miR-96+miR-182 (Figure 2i). Morphologically, these cells appeared to lose membrane protrusions, but did not adhere to their neighboring cells as observed in the miR-200c+miR-141-expressing cells (Figure 2h), consistent with the reduced induction of CDH1 by this cluster.
ZEB1-regulated miRNA clusters block VM. (a) Quantitative reverse transcription–PCR (qRT–PCR) for ZEB1 in MDA-MB-231 cells transfected with siNT control or siZEB1. Error bars represent s.d. of 3 independent experiments. (b) Western blot analysis (left) and quantitation (right) of ZEB1 in control vs knockdown cells. ACTB is loading control. Error bars represent s.d. from 3 independent experiments. (c) Phase images of VM assay in control or ZEB1 knockdown cells. Scale bars=1000 μm. (d) Quantification of total network length over 10 independent experiments. Error bars represent s.d., t-test was performed, ***P<0.001. (e) qRT–PCR for miRNAs in cells transfected for 72 h with siNT or siZEB1. Error bars represent s.d. of 3 independent experiments. T-tests were performed, **P<0.01; ***P<0.001. (f) Phase images of VM assay with NEG or miRNA mimic transfected cells. Scale bars=1000 μm. (g) Quantification of total network length. Error bars represent s.d. for 3 independent experiments. T-tests were performed, *P<0.05. (h) Phase images of MDA-MB-231 cells transfected with miRNAs as shown. Scale bars=100 μm. (i) qRT–PCR following expression of NEG control or miRNA mimics. Error bars represent s.d. of 3 independent experiments. T-tests were performed for each condition vs NEG control, ***P<0.001. (j) Western blot analysis in cells transfected with NEG mimic or miRNA mimics. ACTB is loading control. (k) Quantitation of proteins normalized to ACTB in 3 independent experiments. Error bars represent s.d. T-tests were performed for each protein in cells transfected with each miRNA cluster vs NEG control, *P<0.05, ***P<0.001.
Select miRNAs inhibit VM
To assess a direct role for each of these miRNAs in VM, we transfected the five miRNAs in these two clusters individually into MDA-MB-231 cells, confirmed expression of the miRNAs (Supplementary Figure S3A), and then plated the cells in a VM assay. We found that transient expression of miR-200c, -183, -96 or -182 was capable of blocking VM, while expression of miR-141 did not affect network formation (Figures 3a and b). We confirmed that the loss of network formation was not due to significant differences in cell survival in 0% serum between the control miRNA, miR-141 and other VM-disrupting miRNAs (Supplementary Figure S3B). We tested whether expression of VM-blocking miRNAs affected adhesion of cells to Matrigel or migration in a scratch wound assay. We found that expression of all the VM-blocking miRNAs decreased the rate of adhesion to Matrigel and expression of miR-200c or miR-183 also significantly decreased migration (Supplementary Figures S3C and D). This is consistent with previous reports26, 29, 32, 33 and suggests that the effects of these miRNA clusters on adhesion and migration could contribute to their suppression of VM in mesenchymal breast cancer cells.
Select miRNAs inhibit VM. (a) Representative images of VM in MDA-MB-231 cells transfected with miRNA mimics or NEG. Scale bars=1000 μm. (b) Quantification of total network length. Error bars represent s.d. for at least five independent experiments. T-tests were performed to compare each to NEG control, ***P<0.001. (c) Quantitative reverse transcription–PCR following transfection of MDA-MB-231 cells with miRNA mimics. Error bars represent s.d. from three independent experiments. T-tests were performed, **P<0.01, ***P<0.001. (d) Representative western blot in cells transfected with NEG, miR-200c or miR-141. ACTB is loading control. (e) Quantitation of proteins normalized to ACTB. Error bars represent s.d. of three independent experiments, t-tests were performed to compare to NEG control, **P<0.01, ***P<0.001.
We next took advantage of the fact that miR-200c and miR-141 have very similar seed sequences and many overlapping targets, yet display distinct phenotypic influence on VM when transiently overexpressed. We assessed levels of several known miR-200c targets,34, 35, 36 and found that while both miR-200c and miR-141 target ZEB1 and drive increased expression of CDH1, only miR-200c expression affected the level of the secreted extracellular matrix protein FN1 and the COPII secretory machinery protein SEC23A (Figures 3c–e). As further confirmation that the miR-141 mimic was functional, we also found that it targeted TGFB2 as previously demonstrated27 (Supplementary Figure S3E). Our data indicated that miR-141 targeting of ZEB1 was not sufficient to block VM while loss of ZEB1 expression with siRNA was. Therefore, we also assessed FN1 levels following siZEB1 transfection and found that while miR-141 expression did not affect FN1 levels, siZEB1 significantly decreased FN1 expression (Supplementary Figure S3F). This could be explained by miR-141 having additional targets that function to help maintain FN1 expression or through the possibility that longer-term expression of miR-141 is required to drive FN1 loss. Finally, we cloned the 3′-untranslated region of FN1 into the pMIR-Luc vector and showed, consistent with previous reports36, 37 that miR-200c and miR-96 directly target FN1 (Supplementary Figure S3G).
To determine whether these miRNAs also block VM in an additional cell line, we transiently transfected BT-549 cells with negative control (NEG) or individual miRNA mimics. Once again, we found that miR-200c, -183, -96 and -182 were sufficient to inhibit network formation, while miR-141 was not (Supplementary Figures S3H and I). We assessed the regulation of the miRNA targets in this additional cell line and, similar to the MDA-MB-231 cells, the VM-inhibiting miRNAs drove decreased expression of ZEB1 and FN1, while miR-141 increased FN1 (Supplementary Figure S3J). CDH1 expression was again increased following expression of miR-141 (Supplementary Figure S3J), indicating in both cell lines that re-expression of CDH1 alone is not sufficient to block VM.
COPII-mediated secretion is essential for VM
The specific regulation of FN1 by VM-inhibiting miRNAs points to the importance of FN1 secretion in VM and together with the differential repression of SEC23A by miR-200c and miR-141 led us to ask whether COPII-mediated secretion has a critical role in VM. To test this, we first transfected MDA-MB-231 cells with a SEC23A-targeted siRNA pool or a non-targeting control pool, but despite a 75% knockdown in SEC23A protein levels, we found that siSEC23A cells were still able to undergo VM (Supplementary Figures S4A–D). This implied that SEC23A is not necessary or sufficient for VM, consistent with miR-183, -96 and -182 not affecting its expression but still blocking VM (Figures 3a–c). As there could be functional compensation by other secretory pathway proteins such as SEC23B, for example, that has been shown to partially compensate for loss of SEC23A in development,38 we directly addressed the role of COPII-mediated secretion in VM by transfecting MDA-MB-231 and BT-549 cells with an siRNA pool targeted against SEC13. While SEC13 is not a target of the VM-blocking miRNAs (Supplementary Figure S4E), it is a non-redundant factor in the COPII pathway.39 We confirmed knockdown of SEC13 at the mRNA level in both cell lines (Figure 4a), and found, upon plating these cells in the VM assay, that cells with decreased expression of SEC13 had a significantly reduced ability to form networks (Figures 4b and c).
COPII-mediated secretion is essential for VM. (a) Quantitative reverse transcription–PCR (qRT–PCR) for SEC13 in MDA-MB-231 or BT-549 cells transfected with siNT or siSEC13. Error bars represent s.d. of three (MDA-MB-231) or two (BT-549) independent experiments. (b) Representative images of VM in MDA-MB-231 or BT-549 cells transfected with siNT or siSEC13. Scale bars=1000 μm. (c) Quantification of total network length in VM assays in MDA-MB-231 or BT-549 cells. Error bars represent s.d. for five or three independent experiments, respectively. T-tests were performed to compare each to siNT control, ***P<0.001, *P<0.05. (d) qRT–PCR following expression of a NEG or clustered miRNA mimics. Error bars represent s.d. of three independent experiments. T-tests were performed vs NEG control, ***P<0.001. (h) Western blot for FN1 and SERPINE2 in conditioned media from MDA-MB-231 cells transfected as shown. (f) Quantification of three (FN1) or two (SERPINE2) independent replicates of western blots as shown in e. Error bars represent s.d. T-tests were performed for FN1 quantitation compared to NEG control. **P<0.05; ***P<0.001. (g) qRT–PCR following transfection of MDA-MB-231 cells with miRNA mimics. Error bars represent s.d. from three independent experiments. T-tests were performed, *P<0.05, **P<0.01, *** P<0.001. (h) Western blot for LRP1 following transfection of MDA-MB-231 cells with miRNA mimics. Shown is a representative image of two independent experiments. GAPDH is loading control. (i) qRT–PCR following transfection of MDA-MB-231 cells with miRNA mimics. Error bars represent s.d. from three independent experiments. T-tests were performed, *P<0.05, ***P<0.001.
As COPII-mediated secretion appeared essential for VM and as the secreted protein FN1 was consistently targeted by VM-inhibiting miRNAs, we examined whether other secreted proteins that may contribute to VM were affected by these miRNAs. A recent report implicated secreted protease inhibitors, SerpinE2 and Slpi, as critical for VM in mouse mammary tumor cells10 and other reports have shown that miR-200c targets CXCL1 and interleukin 8 in tumor and endothelial cells to regulate tumor angiogenesis.40 Therefore, we tested whether the ZEB1-regulated miRNAs affected expression of these factors. We found that SERPINE2 was downregulated by expression of both miR-200c+141 and miR-183+96+182 (Figure 4d). SERPINE2 does not appear to have a canonical seed sequence for any of these miRNAs, so it is likely an indirect target. In MDA-MB-231 cells, downregulation of SERPINE2 was mediated by the individual VM-inhibiting miRNAs: miR-200c; -183; -96; and 182, whereas in BT-549 cells, only miR-183 and -96 caused a significant decrease in SERPINE2 expression while miR-141 increased its expression (Supplementary Figure S4F). The fact that miR-200c does not affect SERPINE2 in BT-549 cells could contribute to the more modest effect of miR-200c on VM inhibition in this line (Supplementary Figures S3H and I). We found that miR-200c+141 caused an upregulation of, and miR-183+96+182 had no effect on, SLPI expression (Supplementary Figure S4G). We found no significant change in Interleukin 8 (Supplementary Figure S4H). CXCL1 levels did appear decreased with expression of some of these miRNAs (Supplementary Figure S4H), however, it was equally targeted by miR-200c and miR-141, which differentially affect VM (Figure 3a). Thus, it is unlikely that SLPI, CXCL1 or interleukin 8 significantly contribute to the cell-intrinsic and autocrine signaling regulating VM in this system.
As others have reported global effects on secretion with miR-200c expression,34, 41 we next tested whether secretion was globally suppressed. We did not observe significant changes in the total protein content in conditioned serum-free media following expression of miR-200c, miR-200c+miR-141 or miR-183+miR-96+miR-183 (Supplementary Figure S4I). Western blot analysis, however, revealed that the level of secreted FN1 was greatly decreased (Figures 4e and f). Surprisingly, we observed an increase in SERPINE2 levels in the conditioned media of cells expressing miR-200c despite the fact that the mRNA expression was decreased (Figures 4e and f). This led us to ask whether the expression of cellular receptors to these secreted proteins was also altered with expression of these miRNAs. LRP1 binds to SERPINE2/protease complexes to induce ERK signaling and MMP9 expression, and has been linked to breast cancer metastasis.42 LRP1 has also been shown to be a direct target of miR-200.34 Here we confirmed that its expression was decreased at both the mRNA and protein levels following expression of either miR-200c or miR-183 (Figures 4g and h). The FN1 receptor ITGB1 was also decreased by miR-183, -96 and -182 consistent with previous reports that it is a direct target of this miRNA cluster26, 32 (Figure 4i). Similar regulation of LRP1 and ITGB1 by these miRNAs was also observed in BT-549 cells (Supplementary Figure S4J). Together, these data indicate that COPII-mediated secretion is essential for VM and suggest that miR-200c and the miR-183.96.182 cluster may inhibit VM through regulation of autocrine signaling affecting both secreted factors, including FN1 and SERPINE2, and their receptors.
Increased expression of FN1 and SERPINE2 is regulated by serum withdrawal and is essential for VM
Upon determining that COPII-mediated secretion was essential for VM, we circled back to ask whether the extrinsic condition of serum withdrawal affected the expression of secreted factors to influence this process. We focused on FN1 and SERPINE2, as their expression was regulated by VM-inhibiting miRNAs, and assessed the levels of these factors in four TNBC cell lines in the presence or absence of serum. We found that FN1 and SERPINE2 were expressed at higher baseline levels in the VM-competent MDA-MB-231 and BT-549 lines as compared to MDA-MB-453 and MDA-MB-468 lines, which do not undergo VM (Figure 5a). In addition, we found that withdrawal of serum caused a significant increase in FN1 and SERPINE2 in MDA-MB-231 cells, as well as a significant increase of SERPINE2 in BT-549 cells (Figure 5a). FN1 was expressed at a higher level in BT-549 cells regardless of serum levels (Figure 5a). While expression of FN1 and SERPINE2 increased in the MDA-MB-468 cell line in response to serum withdrawal, the levels still did not reach the baseline level observed in the VM-competent cells (Figure 5a).
Increased expression of FN1 and SERPINE2 is regulated by serum withdrawal and is essential for VM. (a) Quantitative reverse transcription–PCR (qRT–PCR) in breast cancer cell lines cultured with 10 or 0% serum for 48 h. Error bars represent s.d. of three independent experiments. T-tests were performed for each line comparing 0vs10% conditions, **P<0.01, ***P<0.001. (b, d) Representative images of VM in control and FN1 (b) or SERPINE2 (d) knockdown cells. Scale bars=1000 μm. (c, e) Quantification of total network length for VM assays as shown in b and d. Error bars represent s.d. for at five and four independent experiments, respectively. T-tests were performed to compare each to siNT control, **P<0.01. (f) Representative images of VM in control and FN1 or SERPINE2 knockdown in BT-549 cells. Scale bars=1000 μm. (g) Quantification of total network length. Error bars represent s.d. for three independent experiments. T-tests were performed to compare each to siNT control, *P<0.05. (h) Representative phase images of cell morphology in siNT or siSERPINE2 cells. Scale bars=100 μm. (i) qRT–PCR for FN1 and CDH1 in cells transfected with siNT or siSERPINE2. Error bars represent s.d. of three independent experiments. T-test was performed, ***P<0.001. (j) qRT–PCR for FN1 and SERPINE2 in cells transfected with NEG or clustered miRNA mimics for 48 h, then cultured with 10 or 0.5% serum for 48 h. Error bars represent s.d. of three independent experiments. T-tests were performed comparing 0.5vs10% in the control or comparing cells transfected with miRNAs vs NEG in matching serum conditions, ***P<0.001. (k) Western blot for FN1 for cells as in h. ACTB is loading control. (l, m) Quantification of total network length of VM assays from VM-incompetent cells plated in 0% serum (l) or VM-competent cells plated in 5% serum (m) in the presence or absence of recombinant FN1 and SERPINE2 as shown. Error bars represent s.d. of two independent experiments.
To determine whether the increase in FN1 or SERPINE2 was necessary for VM under these conditions, we transfected MDA-MB-231 cells with pooled or multiple individual siRNAs against each of these factors, confirmed knockdown (Supplementary Figures S5A and B), and plated cells in the VM assay. Knockdown of either FN1 or SERPINE2 with either the pooled or individual siRNAs significantly inhibited VM (Figures 5b–e; Supplementary Figure S5C). In addition, we transfected BT-549 cells with siRNA pools to FN1 and SERPINE2, confirmed decreased expression of the mRNAs (Supplementary Figure S5D) and found that knockdown of either of these factors again inhibited VM in this cell line (Figures 5f and g). Importantly, there was no significant increase in cell death when cells were transfected with these siRNAs and plated in 0% serum conditions (Supplementary Figure S5E). Interestingly, we found that knockdown of SERPINE2 resulted in loss of membrane protrusions, decreased expression of FN1 and increased CDH1 expression (Figures 5h and i), suggesting a cell-intrinsic or autocrine function of this secreted factor in regulating the mesenchymal phenotype.
We next tested whether the ZEB1-regulated miRNA clusters affected expression of these secreted factors in response to serum withdrawal. We transfected MDA-MB-231 cells with a control mimic, miR-200c+miR-141 or miR-183+miR-96+miR-182, and then assessed levels of FN1 and SERPINE2 in response to serum withdrawal. As in Figure 5a, we again observed that both FN1 and SERPINE2 were increased upon serum withdrawal, and importantly we found that expression of miR-200c+miR-141 or miR-183+miR-96+miR-183 was able to decrease expression of both FN1 and SERPINE2, such that even upon serum withdrawal, cells expressing these miRNA clusters did not express these factors at levels equivalent to the control at baseline (Figure 5j). This was confirmed for FN1 protein levels by western blot analysis (Figure 5k). Finally, we tested whether FN1 and SERPINE2 were sufficient to induce VM by adding recombinant proteins to the VM-incompetent cell lines MDA-MB-453 and MDA-MB-468 when these cells were plated on Matrigel. We found that the addition of these two secreted proteins was not sufficient to drive these cell lines to undergo VM (Figure 5l). However, addition of these recombinant proteins was sufficient to induce VM in 5% serum in the VM-competent cell lines MDA-MB-231 and BT-549 (Figure 5m), where 5% serum normally blocks this phenotype (Supplementary Figures S1C–F). This is consistent with our model of increased secretion of these factors in serum-free conditions contributing to this process as well as with decreased expression of these factors and their receptors in the VM-incompetent cell lines (Figure 5a; Supplementary Figure S5F).
Expression of miR-200c reduces VM in vivo
To determine whether expression of miR-200c alters VM in vivo, we first generated clonal stable MDA-MB-231 cell lines overexpressing miR-200c or a control vector using a piggyBac system. We confirmed overexpression of miR-200c in this line (Supplementary Figure S6A) and also confirmed mRNA expression changes, including decreased expression of ZEB1, FN1 and SERPINE2 along with increased expression of CDH1 (Figure 6a), similar to what we had observed with transient overexpression (Figure 3c; Supplementary Figure S4F). We then verified that stable expression of miR-200c inhibited the ability of these cells to undergo VM (Figures 6b and c). To determine whether overexpression of miR-200c affected the vascularity of in vivo tumors, we orthotopically implanted these cells into the mammary fat pad of immunocompromised severe combined immunodeficiency (SCID) mice (N=6 tumors each for control and miR-200c-expressing lines) and allowed the tumors to grow for 6 weeks. We assessed tumor growth over time, and found that the miR-200c-expressing cells grew slightly slower than the control cells in vivo (Figure 6d). Six weeks after implantation, tumors were isolated from all mice. We confirmed at the protein level that miR-200c-expressing tumors had increased expression of CDH1 and decreased expression of ZEB1 and FN1 (Figure 6e; Supplementary Figure S6B). SERPINE2 levels, however, were not decreased in the miR-200c-expressing tumor lysates (Figure 6e; Supplementary Figure S6B), which could be due either to stromal cell expression of SERPINE2 within the tumor microenvironment with both control and miR-200c-overexpressing cell lines or to increased expression in the tumor cells under in vivo vs in vitro conditions. We next assessed the tumor histology by hematoxylin and eosin and found a striking increase in necrotic area in the miR-200c-expressing tumors, suggesting an overall decrease in vascularization (Figures 6f and g). It is possible that the decreased vascularity resulting in decreased delivery of oxygen and nutrients contributed to the elevated baseline levels of SERPINE2 in these tumors, similar to the in vitro removal of serum (Figure 5k). Finally, we assessed whether the tumor cells were contributing to vascular lacunae in the tumor tissue. We performed immunohistochemistry (IHC) for CD31 in combination with periodic acid schiff (PAS) staining to assess both PAS+/CD31+ endothelial vessels as well as PAS+/CD31− vessel-like lacunae indicating VM (Figure 6h). We found that the total number of PAS+ lacunae was significantly decreased in the miR-200c-expressing tumors (Figure 6i). During this analysis, we also observed vascular lacunae that were mosaic in nature such that CD31+ and CD31− cells coexisted within a single PAS+ structure (Figure 6h, yellow arrowheads). To determine whether human tumor cells could contribute to vascular lacunae containing CD31+ and CD31− cells, we performed immunofluorescence for CD31 and a human-specific anti-mitochondria antibody. This analysis revealed that CD31-negative tumor cells could coexist with mouse endothelial cells in mosaic vessels (Supplementary Figure S6C). We then assessed our IHC images to determine the number of PAS+ channels that were endothelial vessels (CD31+) or VM containing lacunae (including CD31− and mosaic vessels) and found a significantly decreased number of both types of vessel structures in the miR-200c-expressing tumors (Figures 6j and k). The average number of each type of vessel structure counted in individual tumors is provided in a table in Supplementary Figure S6D. In addition, we found that some tumor cells contributing to vessels could also express CD31 (Supplementary Figure S6C). This is consistent with previous reports of tumor cells displaying plasticity toward an endothelial phenotype and expressing endothelial markers.8, 9, 43, 44
Expression of miR-200c reduces VM in vivo. (a) Quantitative reverse transcription–PCR in stable clonal lines for genes as shown. (b) Representative images of VM in control and miR-200c-expressing stable lines. Scale bars=1000 μm. (c) Quantification of total network length from b. Error bars represent s.d. for three independent experiments. T-tests were performed to compare to control, **P<0.01. (d) Tumor burden over time for orthotopic xenografts of control and miR-200c-expressing clonal cell lines (n=6 tumors each). T-test was performed on area under the curve for each tumor, ***P<0.001. (e) Quantification of western blot analysis (blots are shown in Supplementary Figure S6B) for proteins shown normalized to GAPDH for each of the six control and six miR-200c-expressing tumors. T-tests to compare to control levels were performed, *P<0.05; **P<0.01. (f) × 1.5 images of hematoxylin and eosin sections of two representative control and miR-200c-expressing tumors. (g) Quantitation of percent necrotic area for all tumors. Error bars represent s.d., t-test was performed, ***P<0.001. (h) Representative images of CD31 IHC with PAS staining on sections of control and miR-200c-expressing tumors. Scale bars=100 μm. Arrowheads indicate CD31-positive vessels (red), CD31-negative vascular lacunae (green) or mosaic vessels (yellow). Insets from boxed regions shown below. (i–k) Quantitation of total PAS+ vascular lacunae (i), PAS+/CD31+ vessels (j) or PAS+/CD31− or mosaic vascular lacunae (k) as measured in 16 randomly selected fields from 3 non-adjacent sections of each tumor. Shown is the average number of vascular lacunae per tissue section (total counted on 16 fields) for each of the six tumors in each set. T-test was performed, *P<0.05; **P<0.01.
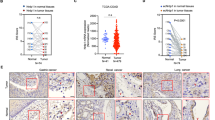
Autocrine signaling factors FN1, ITGB1, SERPINE2 and LRP1 predict poor survival in early-stage breast cancer
Finally, as FN1, SERPINE2, LRP1 and ITGB1 were expressed at higher levels in the mesenchymal cell lines that we tested, we sought to determine whether this was true across a broader panel of cell lines. We assessed the levels of FN1, ITGB1, SERPINE2 and LRP1 in previously published RNA-seq data from a cohort of breast cancer cell lines.45 FN1 and LRP1 were found to be significantly higher in claudin-low as compared to luminal subtype cell lines, while SERPINE2 and ITGB1 levels were significantly higher in both claudin-low and basal-like cell lines as compared to luminal subtype cell lines (Figure 7a; Supplementary Figure S7). Next, we used cBioPortal46, 47 to analyze overall patient survival for groups with high or low expression of these factors in The Cancer Genome Atlas data set.48 We found that in stage I, II and III breast cancer, tumors with low expression (heterozygous loss or homozygous deletion) of MIR200C or MIR183, along with a gain, amplification or increased (>2-fold) expression (‘high’ expression) of FN1 and SERPINE2 had significantly decreased patient survival with a log-rank test P-value of 0.011 and 0.0362, respectively (Figures 7b and c). Increased expression of all of the autocrine pathway genes that we found regulated by these VM-inhibiting miRNAs, FN1, ITGB1, SERPINE2 and LRP1, together also had significantly decreased patient survival with a log-rank test P-value of 0.00133 (Figure 7d). Consistent with our data showing similar regulation of FN1 and SERPINE2, we found that there was a significant tendency for co-occurrence of high FN1 and high SERPINE2 in these tumors with a log odds ratio of >3 (P<0.001). Separating these genes into sets of high FN1 and ITGB1 or high SERPINE2 and LRP1, we found that high expression of either combination could predict decreased survival, with the SERPINE2 and LRP1 combination having the greatest effect with a log-rank test P-value of 6.368e-4 (median months survival for SERPINE2 and LRP1 high=113.7 and for SERPINE2 and LRP1 low=212.09; Figures 7e and f). Together, these data suggest that FN1 and SERPINE2 are upregulated in cancer cells in response to a nutrient-deficient environment, that expression of these factors and their cellular receptors can be kept low by ZEB1-regulated miRNAs, and that high expression of these factors in tumors correlates with decreased overall survival.
Autocrine signaling factors FN1, ITGB1, SERPINE2 and LRP1 predict poor survival in early-stage breast cancer. (a) Plots comparing FN1, ITGB1, SERPINE2 and LRP1 levels in RNA-seq data from subtyped breast cancer cell lines. A one-way analysis of variance with Tukey’s test was performed, *P<0.5, ***P<0.001. (b–f) Survival curves from cBioPortal showing survival of patients with stage I, II or III breast cancer with the following genes altered. Log-rank test P-values are shown. (b) Tumors that had a low level (defined by heterozygous loss or homozygous deletion) of MIR200C along with high expression (defined by a gain, amplification or fold expression >2) of FN1 and SERPINE2 (red) compared to those that did not (blue). (c) Tumors that had a low level of MIR183 along with high expression of FN1 and SERPINE2 (red) compared to those that did not (blue). (d) Tumors that had a high level of FN1, ITGB1, SERPINE2 and LRP1 (red) compared to those that did not (blue). (e) Tumors that had a high level of FN1 and ITGB1 (red) compared to those that did not (blue). (f) Tumors that had a high level of SERPINE2 and LRP1 (red) compared to those that did not (blue).
Discussion
Tumor cells exposed to a hypoxic microenvironment have been shown to respond not only by inducing sprouting angiogenesis but also by directly contributing to tumor vasculature through VM and/or transdifferentiation to an endothelial phenotype. Here we utilized a three-dimensional culture system to study VM and demonstrated that nutrient deprivation induces VM in mesenchymal breast cancer cells. This was accompanied by increased expression of angiogenic and endothelial differentiation gene signatures. Recent evidence suggests that the ability of tumor cells to mimic endothelial phenotypes benefits tumor growth and metastatic progression.7, 9, 10 Understanding the precise mechanisms underlying this process, therefore, will be important to developing effective therapeutics that inhibit vascularization as well as metastasis. We examined multiple TNBC cell lines for their ability to undergo VM in response to serum withdrawal and found that mesenchymal, claudin-low like cells undergo this transition, consistent with these cells having increased vascular endothelial growth factor/hypoxia gene signature expression and endothelial as well as stem-like properties.49, 50 As the capacity to undergo VM appears to be influenced by cell-intrinsic pathways, we identified specific ZEB1-repressed miRNAs, including miR-200c, miR-183, miR-96 and miR-182 that are critical negative regulators of the ability of breast cancer cells to acquire endothelial phenotypes. It is interesting to note that while both of the co-regulated miRNAs miR-200c and miR-141 induce upregulation of E-cadherin, only miR-200c blocks VM. This suggests that it is not the epithelial phenotype itself that is sufficient to block VM, but rather it is likely to be the effect of another target of miR-200c in this system. We also showed that miR-200c can inhibit vascular phenotypes of breast cancer cells in vivo, consistent with another recent report.44
In our effort to understand the molecular regulation of VM, we uncovered a critical role for COPII-mediated secretion in this process and further showed that the secreted proteins FN1 and SERPINE2 have an important role in this response of mesenchymal breast cancer cells to nutrient deficiency. We found that serum withdrawal induces an upregulation of both FN1 and SERPINE2 in mesenchymal breast cancer cells and that expression of the VM-inhibiting miRNAs could inhibit the upregulation of these factors as well as expression of their cellular receptors, ITGB1 and LRP1. FN1 has previously been linked to endothelial function, as its autocrine expression and assembly into a matrix has been shown to be critical for endothelial phenotypes, including new vessel formation.51, 52 Our work suggests that tumor cells that undergo VM also require expression of FN1, as its loss suppressed VM and its addition could facilitate network formation. SERPINE2, as a serine protease inhibitor, is known to inhibit the activity of many proteases, including thrombin, urokinase, plasmin and trypsin. While increased protease activity has been associated with extracellular matrix degradation and subsequent tumor invasion, SERPINE2 has also been found to be upregulated in many cancer types and has been associated with metastasis.53, 54, 55 In pancreatic cancer, overexpression of SERPINE2 was linked to increased extracellular matrix production in tumors,56 and in lung fibroblasts during idiopathic pulmonary fibrosis, SERPINE2 was found to increase FN1 expression.57 In this context, our work indicating that SERPINE2 and FN1 are required for breast cancer VM suggests that they may cooperate to support extracellular matrix development or maturation that allows for endothelial-like vascular network formation.
SerpinE2 was recently described as a critical factor in mouse mammary tumor VM and shown to be expressed in the metastatic tumor cells within a heterogeneous tumor.10 The anticoagulant function of SerpinE2 was linked to increased vessel leakiness leading to increased circulation and eventual metastasis of breast cancer cells.10 While this function of SerpinE2 likely contributes to metastatic capacity, it does not address how the expression of SERPINE2 specifically regulates the cell-autonomous expression of endothelial phenotypes. Here we observed that the loss of SERPINE2 in breast cancer cells affects cell morphology, resulting in loss of membrane protrusions and, consistent with this, SERPINE2 knockdown cells also have decreased FN1 and increased CDH1 expression. It is possible that in addition to a role remodeling the extracellular matrix, SERPINE2 has a role in cell-intrinsic phenotypes to promote metastatic behavior, including VM. A previous report demonstrated that SERPINE2, when bound to a protease, interacts with the LRP1 receptor for internalization, resulting in upregulation of ERK activity.42 LRP1 has also been shown to be a target of miR-200c,34 and we show here that its expression is regulated by VM-inhibiting miRNAs in our system. In light of these results, we hypothesize that SERPINE2 signals in an autocrine manner through LRP1 to contribute to cellular signaling pathways supporting VM-related phenotypes.
Together, our results suggest that mesenchymal cancer cells, with low expression of ZEB1-regulated miRNA clusters, respond to a nutrient-deficient microenvironment by adopting endothelial phenotypes. We show that nutrient deficiency leads to an upregulation of FN1 and SERPINE2, and that these factors are critical for VM. Finally, we show that the autocrine signaling factors that we identified to be regulated by ZEB1-repressed miRNAs and to functionally contribute to VM are also correlated with decreased patient survival. These new mechanistic insights could contribute to new strategies to prevent the tumor cell plasticity toward endothelial phenotypes that supports tumor vascularization and metastasis.
Materials and methods
Cell culture and transfection
MDA-MB-231, BT-549, Hs578T, MDA-MB-468, MDA-MB-453, HCC1395 and HEK293 cells were purchased from American Type Culture Collection (Manassas, VA, USA); SUM159PT were obtained from Dr Joe Gray (Oregon Health & Science University; OHSU); and HUVECs were purchased from EMD-Millipore (Billerica, MA, USA). All cell lines were validated by short tandem repeat profiling and confirmed to be free of mycoplasma contamination. MDA-MB-231, Hs578T, MDA-MB-468, MDA-MB-453 and HEK293 cells were grown in high-glucose Dulbecco’s modified Eagle’s medium (Gibco, Thermo Fisher, Waltham, MA, USA) supplemented with 10% fetal bovine serum (FBS; Atlanta Biologicals, Flowery Branch, GA, USA). BT-549 and HCC1395 cells were grown in RPMI (Gibco) supplemented with 10% FBS. SUM159PT were grown in Hams-F12 media (Gibco) supplemented with 5% FBS, 10 mm HEPES, 4 μg/ml insulin and 0.5 μg/ml hydrocortisone. HUVECs were grown in complete EndoGRO medium (Millipore).
MiRIDIAN miRNA mimics as well as ON-TARGETplus SMARTpool and ON-TARGETplus individual siRNAs (listed in Supplementary Table 2) were purchased from GE Dharmacon (Lafayette, CO, USA), and transfected using Lipofectamine 2000 or Lipofectamine 3000 (Life Technologies) according to the manufacturer’s protocol at a final concentration of 25 nm. BT-549 cells were transfected with LF2000 on 2 consecutive days to increase transfection efficiency. Phase images of cells were taken on an EVOS-FL scope (Thermo Fisher).
Quantitative reverse transcription–PCR
RNA was isolated using Trizol (Life Technologies) according to the manufacturer’s instructions. For mRNA analysis, RNA was treated with DNAse I (Roche, Basel, Switzerland) and purified over a Qiagen (Hilden, Germany) RNeasy column. First strand synthesis was performed using a High Capacity cDNA Reverse Transcription kit (Life Technologies). Quantitative PCR was performed using SYBR green (Life Technologies) on a Step One Real Time PCR system with Step One software (Life Technologies). All primer sequences are provided in Supplementary Table 3. TBP was used for normalization. For miRNA analysis, RNA was used in Taqman quantitative reverse transcription–PCR assays according to the manufacturer’s protocol (Life Technologies). U6 snRNA was used for normalization.
Stable cell lines
The genomic region containing hsa-miR-200c.141 was cloned from genomic DNA of MDA-MB-453 cells into the pGEM-Teasy vector system (Promega, Madison, WI, USA). Primers are listed in Supplementary Table 3. The cloned region was sequenced and then digested with EcoRI and PvuII. This 931 bp band was further digested with SalI to generate a 589 bp band containing only miR-200c. This band and the NotI-digested piggybac vector, PB-CAG-GFP-iNeo, were blunted with Klenow (New England Biolabs, Ipswich, MA, USA) and ligated to generate PB-CAG-GFP-hsa-miR200c-iNeo plasmid with miR-200c cloned downstream of green fluorescent protein (GFP) but before the polyA sequence. MDA-MB-231 cells were transfected with a 1:3 ratio of the pCAG-PBase transposase plasmid58 to the PB-CAG-GFP-hsa-miR200c-iNeo or control plasmid with Lipofectamine 2000. Clonal cell lines were selected in 2 mg/ml G418 and expression of miRNAs and targets were confirmed with quantitative reverse transcription–PCR.
VM assay
Cells were trypsinized, counted, washed to remove serum, and plated onto a thick (40 μl) layer of growth factor reduced Matrigel (BD Biosciences) in a 96-well plate. Cells were resuspended in 150 μl of serum-free Dulbecco’s modified Eagle’s medium for a total of 1.5 × 104 cells/well. Wells were imaged 18–24 h later at × 4 on EVOS-FL scope for the presence or absence of networks. Total network length was calculated with the neurite tracing function in the NeuronGrowth plug-in in ImageJ.59 For co-culture assays, breast cancer cells and HUVECs were incubated for 30 min before trypsinization in serum-free media containing 1 μm Cell Tracker Green CMFDA or Cell Tracker Orange CMRA (Life Technologies), respectively. Cells were then plated on Matrigel in EndoGRO media (2% FBS) at a concentration of 1 × 104 HUVECs+0.5 × 104 cancer cells/well. Recombinant FN1 (1918-FN-02M, R&D Systems, Minneapolis, MN, USA) and SERPINE2 (2980-PI-010, R&D Systems) were added to VM assays at concentrations of 1 and 10 μg/ml, respectively. All VM experiments were repeated in multiple independent experiments as indicated in figure legends. Representative images are shown.
Flow cytometry
Cells were stained with APC α-CD44 (G44-26, BD Pharmingen, Franklin Lakes, NJ, USA) and PE α-CD24 (ML5, BD Pharmingen) at 1:10 dilutions in FACS Buffer (1% bovine serum albumin in phosphate-buffered saline). Flow cytometry was performed on a Canto II (BD Biosciences) with FACS Diva software (BD Biosciences) and data were analyzed using FlowJo software (FlowJo LLC, Ashland, OR, USA). Experiments were repeated in three independent biological experiments, and representative results are shown.
RNA-seq and Gene Set Enrichment Analysis analysis
For RNA-seq, cells plated 24 h on Matrigel were dissociated with Dispase (BD Biosciences), total RNA was isolated with Trizol and sequencing libraries constructed using a TruSeq RNAseq kit (Illumina, San Diego, CA, USA). Briefly, poly(A)+ RNA was isolated from 1μg of total RNA (per sample) using oligo-dT-coated magnetic beads. The recovered RNA was then chemically fragmented. First strand cDNA was generated using random hexamers as primers for reverse transcriptase. Following second strand synthesis, the ends of the double-stranded fragments were repaired and then a single ‘A’ nucleotide was added to each end. Illumina adaptors were ligated to the cDNAs. Limited-round (11-cycle) PCR was used to amplify the material to yield the final libraries. Library concentration was determined using real-time PCR with primers complementary to the Illumina adaptors. Sample libraries were diluted and applied to an Illumina single-read flow cell at a concentration appropriate to generate about 180 million reads per lane. All libraries were prepared with indexing barcodes to permit multiplexing in a single lane. The 100-cycle single-read sequencing was used to generate base call files. Illumina’s CASAVA package was used to assemble the reads into standard fastq formatted data.
RNA-seq analysis was performed as described previously.60 Briefly, data were aligned using Bowtie version 1.0.0 (http://bowtie.cbcb.umd.edu) to human genome version hg19, and custom R scripts and software were used to count tags that aligned to the exons of University of California, Santa Cruz RefSeq gene models. The χ2-statistic was calculated for each gene model and the Benjamini–Hochberg false discovery rate adjustment was applied. RNA-seq data can be accessed at the Gene Expression Omnibus, accession # GSE77179. Each sample was normalized based on the total reads that aligned to exons to calculate fold change and expression difference between classes. For comparison to mouse genes upregulated in Flk1hi differentiating embryonic stem cells after expression of Snail, microarray data from GSE26524 was used to identify genes upregulated in Flk1hi cells sorted from differentiating embryonic stem cells in DKK1+dox condition (dox-induced Snail expression) compared to the genes expressed with DKK1 treatment alone. Genes were sorted by fold change, adjusted to represent human gene symbols, duplicated genes removed and the top 350 genes, all at least 4.5-fold enriched, were included in the comparison. This list is included as Supplementary Table 4. This gene signature, as well as others in the Molecular Signatures Database as indicated, was compared to the RNA-seq data of breast cancer cells plated on Matrigel in the presence of 10 or 0% serum using the Gene Set Enrichment Analysis software.18
Western blots
Cells were lysed in AB lysis buffer and western blot analysis was performed. Blots were visualized and bands quantified on an Odyssey imaging system (LI-COR Biosciences, Lincoln, NE, USA) as described.61 Primary antibodies used were ZEB1 (H-102, Santa Cruz Biotechnology, Santa Cruz, CA, USA, used at a 1:200 dilution), SEC23A (NBP1-32773, Novus Biologicals, Littleton, CO, USA, used at a 1:5000 dilution), CDH1 (36/E-cadherin, BD Biosciences, used at a 1:5000 dilution), FN1 (C-20, Santa Cruz Biotechnology, used at a 1:100 or 1:200 dilution), SERPINE2 (8C4.1, Millipore, used at 1:1000), LRP1 (EPR3724, Abcam, Cambridge, UK, used at 1:5000), GAPDH (6C5, Thermo Fisher, used at 1:10 000) and ACTB (AC-15, Sigma-Aldrich, St. Louis, MO, USA, used at a 1:10 000 dilution). Secondary antibodies were anti-Rabbit (Alexa Fluor 680), anti-Rabbit (Alexa Fluor 800), anti-Goat (Alexa Fluor 680), anti-Mouse (Alexa Fluor 680) or anti-Mouse (Alexa Fluor 800) (Molecular Probes, Eugene, OR, USA), used at a 1:10 000 dilution.
Adhesion and migration assays
Adhesion assays were performed on 96-well plates coated with 2.5 μg/well Matrigel and blocked with heat-inactivated 1% bovine serum albumin for 30 min at 37 °C. MDA-MB-231 cells transfected for 48 h with miRNA mimics were counted and plated in serum-free media at 3 × 104 cells/well for 45 min, washed once with serum-free media, fixed in 2% paraformaldehyde, stained with 0.5% crystal violet in 20% MeOH and then washed with ddH2O. Stain was eluted in methanol and absorbance at 600 nm for each well was determined. For migration assays, MDA-MB-231 cells transfected for 48 h with miRNA mimics were counted and plated onto image lock 96-well plates (Essen Biosciences, Ann Arbor, MI, USA) at a density of 40 000 cells/well. Scratch wounds were generated with an IncuCyte WoundMaker (Essen Biosciences) and images acquired on IncuCyte Zoom (Essen Biosciences) every 4 h. IncuCyte software was used to mask the original wound area and measure confluence of cells within that area over 3 days. Area under the curve was calculated for at least three independent experiments, and t-tests of these values performed to determine significance.
Death assay
MDA-MB-231 cells transfected for 48 h with miRNA mimics were counted, and 10 000 cells/well were plated into 96-well plates in serum-free media. After 24 h, CellTox Green Dye (Promega G873B, used at 1:2000) was added to detect dead cells and images were acquired using an IncuCyte Zoom. Total cell count per well was then determined by adding Nuclear-ID Red DNA stain (Enzo Life Sciences, Farmingdale, NY, USA, ENZ-52406, used at 1:1000) and again acquiring images on an IncuCyte Zoom. Percent dead cells was calculated as the ratio of CellTox Green-positive to Nuclear-ID Red DNA-positive cells.
Conditioned media analysis
MDA-MB-231 cells were plated in six-well plates at 1 × 105 cells/well, and were transfected with miRNA mimics the next day. Forty-eight hours after transfection, cells were washed four times with serum-free media and then grown for an additional 24 h in 1.5 ml serum-free Dulbecco’s modified Eagle’s medium. Media was then collected and spun at 500 × g to remove cellular debris. Conditioned media was then concentrated over Amicon Ultra centrifugal filers (Ultracel-10K, Millipore) and run on a 4–12% gradient SDS–polyacrylamide gel electrophoresis gel. Coomassie staining was performed as described.62, 63 Briefly, gels were fixed for 1 h in 12% (w/v) TCA, stained overnight in 0.12% (w/v) CBB G-250 in 10% (w/v) ammonium sulfate, 10% (v/v) phosphoric acid and 20% (v/v) methanol, and then washed for 1.5 h with destaining solution (25% methanol). Alternatively, proteins were transferred for western blot analysis of FN1 or SERPINE2 as described above.
Luciferase assay
The 3′-untranslated region of human FN1 was cloned from genomic DNA of MDA-MB-453 cells into the pGEM-Teasy vector (Promega) using primers as indicated in Supplementary Table 3 and the sequence was verified. This region was then subcloned into the pMIR-REPORT Luciferase vector (Applied Biosystems, Thermo Fisher). HEK293 cells were co-transfected with this construct, pCMV-β-Gal and individual miRNA mimics (or negative control) using Lipofectamine 2000. Luciferase activity was assessed with Promega luciferase assay kit and a Berthold (Bundoora, VIC, Australia) luminometer and adjusted for β-Gal.
Xenografts and tumor analysis
All animal studies were performed in accordance with the research guidelines for the use of laboratory animals adopted by the OHSU IACUC. On the basis of prior knowledge from these or similar experimental systems, six tumors per group were chosen to satisfy 95% confidence intervals. Balb/c SCID female mice (CBySmn.CB17-Prkdcscid/J, Jackson Laboratory, Bar Harbor, ME, USA) were orthotopically injected in 50% Matrigel into the fourth inguinal mammary fat pad at 7 weeks of age with 4 × 105 MDA-MB 231 cells with stable overexpression of GFP or miR-200c. Tumor burden was determined three times per week through caliper measurements (volume=(W2 × L)/2). Forty-one days post xenograft the mice were perfused with 20 ml PBS with 0.05% heparin, and tumors were collected. Small tumor pieces were flash-frozen and protein extracted by mincing with a Tissue-Tearor (model 985370-395, Biospec Products, Bartletsville, OK, USA) in AB lysis buffer. Westerns were performed as described above. The majority of the tumor was fixed in neutral buffered formalin, paraffin-embedded and sectioned, and hematoxylin and eosin stained by the OHSU Histopathology Shared Resource.
IHC was performed by the OHSU Histopathology Shared Resource on a Ventana Benchmark XT (Roche). Antigen retrieval was performed with Ventana Cell Conditioning Solution 1 for 30 min at 95 °C, anti-CD31 antibody (ab28364, Abcam, used at 1:100 dilution) applied for 30 min at 37 °C and signal detected with the Ventana ultraView Universal DAB Detection kit used with the ImmPRESS HRP anti-rabbit antibody (Vector Laboratories, Burlingame, CA, USA, MP-7401). Slides were then counterstained with PAS. Hematoxylin and eosin as well as IHC and PAS staining was imaged on an Aperio digital slide scanner (Leica Biosystems, Wetzlar, Germany) and viewed on ImageScope. For hematoxylin and eosin tissues, × 1.5 images were then analyzed on ImageJ to quantify the percent necrotic area of the tumor. For IHC analysis, 16 randomly selected × 20 images were obtained for each of three non-adjacent sections for every tumor. These 48 images/tumor were blinded and the number of PAS+ lacunae, PAS+/CD31+, and PAS+/CD31− or CD31 mosaic vascular lacunae was counted. Average number of vessel structures per section is presented for individual tumors. Immunofluorescence was performed on tissue sections using standard protocols with anti-CD31 (ab28364, Abcam, 1:50 dilution) and anti-Mitochondria (ab92824, Abcam, 1:1000 dilution), as well as 4,6-diamidino-2-phenylindole nuclear counterstain. Images were acquired on a Zeiss Axio Scan and analyzed with Zeiss Zen software (Zeiss Microscopy, Thornwood, NY, USA).
Statistical tests
Statistical analysis was performed with GraphPad Prism Software (v5, GraphPad Software, Inc., San Diego, CA, USA). All error bars displayed represent s.d. to show variance between independent experiments or between samples in each group. All t-tests performed were two-tailed tests and results are given as P-values where * indicates P<0.05, ** indicates P<0.01 and *** indicates P<0.001.
References
Weis SM, Cheresh DA . Tumor angiogenesis: molecular pathways and therapeutic targets. Nat Med 2011; 17: 1359–1370.
Folkman J . Role of angiogenesis in tumor growth and metastasis. Semin Oncol 2002; 29 (6 Suppl 16): 15–18.
Quesada AR, Medina MA, Munoz-Chapuli R, Ponce AL . Do not say ever never more: the ins and outs of antiangiogenic therapies. Curr Pharm Des 2010; 16: 3932–3957.
Yang MH, Wu MZ, Chiou SH, Chen PM, Chang SY, Liu CJ et al. Direct regulation of TWIST by HIF-1alpha promotes metastasis. Nat Cell Biol 2008; 10: 295–305.
Conley SJ, Gheordunescu E, Kakarala P, Newman B, Korkaya H, Heath AN et al. Antiangiogenic agents increase breast cancer stem cells via the generation of tumor hypoxia. Proc Natl Acad Sci USA 2012; 109: 2784–2789.
Zhang L, Huang G, Li X, Zhang Y, Jiang Y, Shen J et al. Hypoxia induces epithelial-mesenchymal transition via activation of SNAI1 by hypoxia-inducible factor -1alpha in hepatocellular carcinoma. BMC Cancer 2013; 13: 108.
Xu Y, Li Q, Li XY, Yang QY, Xu WW, Liu GL . Short-term anti-vascular endothelial growth factor treatment elicits vasculogenic mimicry formation of tumors to accelerate metastasis. J Exp Clin Cancer Res 2012; 31: 16.
Soda Y, Marumoto T, Friedmann-Morvinski D, Soda M, Liu F, Michiue H et al. Transdifferentiation of glioblastoma cells into vascular endothelial cells. Proc Natl Acad Sci USA 2011; 108: 4274–4280.
Ricci-Vitiani L, Pallini R, Biffoni M, Todaro M, Invernici G, Cenci T et al. Tumour vascularization via endothelial differentiation of glioblastoma stem-like cells. Nature 2010; 468: 824–828.
Wagenblast E, Soto M, Gutierrez-Angel S, Hartl CA, Gable AL, Maceli AR et al. A model of breast cancer heterogeneity reveals vascular mimicry as a driver of metastasis. Nature 2015; 520: 358–362.
Karroum A, Mirshahi P, Faussat AM, Therwath A, Mirshahi M, Hatmi M . Tubular network formation by adriamycin-resistant MCF-7 breast cancer cells is closely linked to MMP-9 and VEGFR-2/VEGFR-3 over-expressions. Eur J Pharmacol 2012; 685: 1–7.
Francescone RA 3rd, Faibish M, Shao R . A Matrigel-based tube formation assay to assess the vasculogenic activity of tumor cells. J Vis Exp 2011; 55.
Liu TJ, Sun BC, Zhao XL, Zhao XM, Sun T, Gu Q et al. CD133+ cells with cancer stem cell characteristics associates with vasculogenic mimicry in triple-negative breast cancer. Oncogene 2013; 32: 544–553.
Plantamura I, Casalini P, Dugnani E, Sasso M, D'Ippolito E, Tortoreto M et al. PDGFRbeta and FGFR2 mediate endothelial cell differentiation capability of triple negative breast carcinoma cells. Mol Oncol 2014; 8: 968–981.
Lehmann BD, Bauer JA, Chen X, Sanders ME, Chakravarthy AB, Shyr Y et al. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J Clin Invest 2011; 121: 2750–2767.
Herschkowitz JI, Simin K, Weigman VJ, Mikaelian I, Usary J, Hu Z et al. Identification of conserved gene expression features between murine mammary carcinoma models and human breast tumors. Genome Biol 2007; 8: R76.
Chang YS, di Tomaso E, McDonald DM, Jones R, Jain RK, Munn LL . Mosaic blood vessels in tumors: frequency of cancer cells in contact with flowing blood. Proc Natl Acad Sci USA 2000; 97: 14608–14613.
Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA 2005; 102: 15545–15550.
Schoenfeld J, Lessan K, Johnson NA, Charnock-Jones DS, Evans A, Vourvouhaki E et al. Bioinformatic analysis of primary endothelial cell gene array data illustrated by the analysis of transcriptome changes in endothelial cells exposed to VEGF-A and PlGF. Angiogenesis 2004; 7: 143–156.
Manalo DJ, Rowan A, Lavoie T, Natarajan L, Kelly BD, Ye SQ et al. Transcriptional regulation of vascular endothelial cell responses to hypoxia by HIF-1. Blood 2005; 105: 659–669.
Qi J, Nakayama K, Cardiff RD, Borowsky AD, Kaul K, Williams R et al. Siah2-dependent concerted activity of HIF and FoxA2 regulates formation of neuroendocrine phenotype and neuroendocrine prostate tumors. Cancer Cell 2010; 18: 23–38.
Fardin P, Barla A, Mosci S, Rosasco L, Verri A, Versteeg R et al. A biology-driven approach identifies the hypoxia gene signature as a predictor of the outcome of neuroblastoma patients. Mol Cancer 2010; 9: 185.
Mense SM, Sengupta A, Zhou M, Lan C, Bentsman G, Volsky DJ et al. Gene expression profiling reveals the profound upregulation of hypoxia-responsive genes in primary human astrocytes. Physiol Genomics 2006; 25: 435–449.
Gill JG, Langer EM, Lindsley RC, Cai M, Murphy TL, Murphy KM . Snail promotes the cell-autonomous generation of Flk1(+) endothelial cells through the repression of the microRNA-200 family. Stem Cells Dev 2012; 21: 167–176.
Gill JG, Langer EM, Lindsley RC, Cai M, Murphy TL, Kyba M et al. Snail and the microRNA-200 family act in opposition to regulate epithelial-to-mesenchymal transition and germ layer fate restriction in differentiating ESCs. Stem Cells 2011; 29: 764–776.
Li XL, Hara T, Choi Y, Subramanian M, Francis P, Bilke S et al. A p21-ZEB1 complex inhibits epithelial-mesenchymal transition through the microRNA 183-96-182 cluster. Mol Cell Biol 2014; 34: 533–550.
Burk U, Schubert J, Wellner U, Schmalhofer O, Vincan E, Spaderna S et al. A reciprocal repression between ZEB1 and members of the miR-200 family promotes EMT and invasion in cancer cells. EMBO Rep 2008; 9: 582–589.
Gregory PA, Bert AG, Paterson EL, Barry SC, Tsykin A, Farshid G et al. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol 2008; 10: 593–601.
Park SM, Gaur AB, Lengyel E, Peter ME . The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev 2008; 22: 894–907.
Chang CJ, Chao CH, Xia W, Yang JY, Xiong Y, Li CW et al. p53 regulates epithelial-mesenchymal transition and stem cell properties through modulating miRNAs. Nat Cell Biol 2011; 13: 317–323.
Shimono Y, Zabala M, Cho RW, Lobo N, Dalerba P, Qian D et al. Downregulation of miRNA-200c links breast cancer stem cells with normal stem cells. Cell 2009; 138: 592–603.
Li G, Luna C, Qiu J, Epstein DL, Gonzalez P . Targeting of integrin beta1 and kinesin 2alpha by microRNA 183. J Biol Chem 2010; 285: 5461–5471.
Wong CM, Wei L, Au SL, Fan DN, Zhou Y, Tsang FH et al. MiR-200b/200c/429 subfamily negatively regulates Rho/ROCK signaling pathway to suppress hepatocellular carcinoma metastasis. Oncotarget 2015; 6: 13658–13670.
Korpal M, Ell BJ, Buffa FM, Ibrahim T, Blanco MA, Celia-Terrassa T et al. Direct targeting of Sec23a by miR-200 s influences cancer cell secretome and promotes metastatic colonization. Nat Med 2011; 17: 1101–1108.
Choi YC, Yoon S, Jeong Y, Yoon J, Baek K . Regulation of vascular endothelial growth factor signaling by miR-200b. Mol Cells 2011; 32: 77–82.
Howe EN, Cochrane DR, Richer JK . Targets of miR-200c mediate suppression of cell motility and anoikis resistance. Breast Cancer Res 2011; 13: R45.
Jalvy-Delvaille S, Maurel M, Majo V, Pierre N, Chabas S, Combe C et al. Molecular basis of differential target regulation by miR-96 and miR-182: the Glypican-3 as a model. Nucleic Acids Res 2012; 40: 1356–1365.
Zhu M, Tao J, Vasievich MP, Wei W, Zhu G, Khoriaty RN et al. Neural tube opening and abnormal extraembryonic membrane development in SEC23A deficient mice. Sci Rep 2015; 5: 15471.
Ong YS, Tang BL, Loo LS, Hong W . p125A exists as part of the mammalian Sec13/Sec31 COPII subcomplex to facilitate ER-Golgi transport. J Cell Biol 2010; 190: 331–345.
Pecot CV, Rupaimoole R, Yang D, Akbani R, Ivan C, Lu C et al. Tumour angiogenesis regulation by the miR-200 family. Nat Commun 2013; 4: 2427.
Schliekelman MJ, Gibbons DL, Faca VM, Creighton CJ, Rizvi ZH, Zhang Q et al. Targets of the tumor suppressor miR-200 in regulation of the epithelial-mesenchymal transition in cancer. Cancer Res 2011; 71: 7670–7682.
Fayard B, Bianchi F, Dey J, Moreno E, Djaffer S, Hynes NE et al. The serine protease inhibitor protease nexin-1 controls mammary cancer metastasis through LRP-1-mediated MMP-9 expression. Cancer Res 2009; 69: 5690–5698.
Wang R, Chadalavada K, Wilshire J, Kowalik U, Hovinga KE, Geber A et al. Glioblastoma stem-like cells give rise to tumour endothelium. Nature 2010; 468: 829–833.
D'Ippolito E, Plantamura I, Bongiovanni L, Casalini P, Baroni S, Piovan C et al. miR-9 and miR-200 regulate PDGFRbeta-mediated endothelial differentiation of tumor cells in triple-negative breast cancer. Cancer Res 2016; 76: 5562–5572.
Heiser LM, Sadanandam A, Kuo WL, Benz SC, Goldstein TC, Ng S et al. Subtype and pathway specific responses to anticancer compounds in breast cancer. Proc Natl Acad Sci USA 2012; 109: 2724–2729.
Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2012; 2: 401–404.
Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 2013; 6: pl1.
Ciriello G, Gatza ML, Beck AH, Wilkerson MD, Rhie SK, Pastore A et al. Comprehensive molecular portraits of invasive lobular breast cancer. Cell 2015; 163: 506–519.
Harrell JC, Pfefferle AD, Zalles N, Prat A, Fan C, Khramtsov A et al. Endothelial-like properties of claudin-low breast cancer cells promote tumor vascular permeability and metastasis. Clin Exp Metastasis 2014; 31: 33–45.
Prat A, Parker JS, Karginova O, Fan C, Livasy C, Herschkowitz JI et al. Phenotypic and molecular characterization of the claudin-low intrinsic subtype of breast cancer. Breast Cancer Res 2010; 12: R68.
Cseh B, Fernandez-Sauze S, Grall D, Schaub S, Doma E, Van Obberghen-Schilling E . Autocrine fibronectin directs matrix assembly and crosstalk between cell-matrix and cell-cell adhesion in vascular endothelial cells. J Cell Sci 2010; 123 (Pt 22): 3989–3999.
Zhou X, Rowe RG, Hiraoka N, George JP, Wirtz D, Mosher DF et al. Fibronectin fibrillogenesis regulates three-dimensional neovessel formation. Genes Dev 2008; 22: 1231–1243.
Candia BJ, Hines WC, Heaphy CM, Griffith JK, Orlando RA . Protease nexin-1 expression is altered in human breast cancer. Cancer Cell Int 2006; 6: 16.
Nagahara A, Nakayama M, Oka D, Tsuchiya M, Kawashima A, Mukai M et al. SERPINE2 is a possible candidate promotor for lymph node metastasis in testicular cancer. Biochem Biophys Res Commun 2010; 391: 1641–1646.
Wang K, Wang B, Xing AY, Xu KS, Li GX, Yu ZH . Prognostic significance of SERPINE2 in gastric cancer and its biological function in SGC7901 cells. J Cancer Res Clin Oncol 2015; 141: 805–812.
Buchholz M, Biebl A, Neesse A, Wagner M, Iwamura T, Leder G et al. SERPINE2 (protease nexin I) promotes extracellular matrix production and local invasion of pancreatic tumors in vivo. Cancer Res 2003; 63: 4945–4951.
Francois D, Venisse L, Marchal-Somme J, Jandrot-Perrus M, Crestani B, Arocas V et al. Increased expression of protease nexin-1 in fibroblasts during idiopathic pulmonary fibrosis regulates thrombin activity and fibronectin expression. Lab Invest 2014; 94: 1237–1246.
Xu C, Qi X, Du X, Zou H, Gao F, Feng T et al. piggyBac mediates efficient in vivo CRISPR library screening for tumorigenesis in mice. Proc Natl Acad Sci USA 2017; 114: 722–727.
Fanti Z, Martinez-Perez ME, De-Miguel FF . NeuronGrowth, a software for automatic quantification of neurite and filopodial dynamics from time-lapse sequences of digital images. Dev Neurobiol 2011; 71: 870–881.
Farrell AS, Pelz C, Wang X, Daniel CJ, Wang Z, Su Y et al. Pin1 regulates the dynamics of c-Myc DNA binding to facilitate target gene regulation and oncogenesis. Mol Cell Biol 2013; 33: 2930–2949.
Arnold HK, Zhang X, Daniel CJ, Tibbitts D, Escamilla-Powers J, Farrell A et al. The Axin1 scaffold protein promotes formation of a degradation complex for c-Myc. EMBO J 2009; 28: 500–512.
Candiano G, Bruschi M, Musante L, Santucci L, Ghiggeri GM, Carnemolla B et al. Blue silver: a very sensitive colloidal Coomassie G-250 staining for proteome analysis. Electrophoresis 2004; 25: 1327–1333.
Dyballa N, Metzger S . Fast and sensitive coomassie staining in quantitative proteomics. Methods Mol Biol 2012; 893: 47–59.
Acknowledgements
This study was supported by a postdoctoral fellowship from the American Cancer Society 118795-PF-10-022-01-CSM (EML), as well as philanthropic gifts from the Colson Family Foundation and the Prospect Creek Foundation, and R01s from the NCI CA100855 and CA196228 (RCS). Flow Cytometry was performed in the Oregon Health & Science University (OHSU) Flow Cytometry Shared Resource, hematoxylin and eosin and PAS staining as well as CD31 IHC was performed by the OHSU Histopathology Shared Resource, and short read sequencing assays (RNA-seq) were performed by the OHSU Massively Parallel Sequencing Shared Resource. These OHSU Shared Resources are supported by the Knight NCI Cancer Center Support Grant 5P30CA069533. We thank Dr Sen Wu for providing the PB-CAG-GFP-iNeo plasmid. We thank Dr Joe Gray for providing cell lines supported by the National Institutes of Health grant U54 HG008100, National Cancer Institute grant 5P30CA069533 and by the Susan G Komen Foundation SAC110. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health nor does it reflect the position or the policy of the Government, and no official endorsement should be inferred.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Oncogene website
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/4.0/
About this article
Cite this article
Langer, E., Kendsersky, N., Daniel, C. et al. ZEB1-repressed microRNAs inhibit autocrine signaling that promotes vascular mimicry of breast cancer cells. Oncogene 37, 1005–1019 (2018). https://doi.org/10.1038/onc.2017.356
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/onc.2017.356
This article is cited by
-
Functional consequences of A-to-I editing of miR-379 in prostate cancer cells
Scientific Reports (2023)
-
USP22 upregulates ZEB1-mediated VEGFA transcription in hepatocellular carcinoma
Cell Death & Disease (2023)
-
Therapeutic silencing of mTOR by systemically administered siRNA-loaded neutral liposomal nanoparticles inhibits DMBA-induced mammary carcinogenesis
British Journal of Cancer (2022)
-
Physicochemical aspects of the tumour microenvironment as drivers of vasculogenic mimicry
Cancer and Metastasis Reviews (2022)
-
HNRNPD interacts with ZHX2 regulating the vasculogenic mimicry formation of glioma cells via linc00707/miR-651-3p/SP2 axis
Cell Death & Disease (2021)