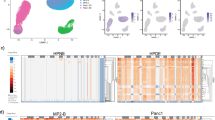
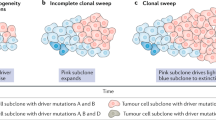
Abstract
Tumor complexity and intratumor heterogeneity contribute to subclonal diversity. Despite advances in next-generation sequencing (NGS) and bioinformatics, detecting rare mutations in primary tumors and metastases contributing to subclonal diversity is a challenge for precision genomics. Here, in order to identify rare mutations, we adapted a recently described epithelial reprograming assay for short-term propagation of epithelial cells from primary and metastatic tumors. Using this approach, we expanded minor clones and obtained epithelial cell-specific DNA/RNA for quantitative NGS analysis. Comparative Ampliseq Comprehensive Cancer Panel sequence analyses were performed on DNA from unprocessed breast tumor and tumor cells propagated from the same tumor. We identified previously uncharacterized mutations present only in the cultured tumor cells, a subset of which has been reported in brain metastatic but not primary breast tumors. In addition, whole-genome sequencing identified mutations enriched in liver metastases of various cancers, including Notch pathway mutations/chromosomal inversions in 5/5 liver metastases, irrespective of cancer types. Mutations/rearrangements in FHIT, involved in purine metabolism, were detected in 4/5 liver metastases, and the same four liver metastases shared mutations in 32 genes, including mutations of different HLA-DR family members affecting OX40 signaling pathway, which could impact the immune response to metastatic cells. Pathway analyses of all mutated genes in liver metastases showed aberrant tumor necrosis factor and transforming growth factor signaling in metastatic cells. Epigenetic regulators including KMT2C/MLL3 and ARID1B, which are mutated in >50% of hepatocellular carcinomas, were also mutated in liver metastases. Thus, irrespective of cancer types, organ-specific metastases may share common genomic aberrations. Since recent studies show independent evolution of primary tumors and metastases and in most cases mutation burden is higher in metastases than primary tumors, the method described here may allow early detection of subclonal somatic alterations associated with metastatic progression and potentially identify therapeutically actionable, metastasis-specific genomic aberrations.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Prasad V, Fojo T, Brada M . Precision oncology: origins, optimism, and potential. Lancet Oncol 2016; 17: e81–e86.
Collins DC, Sundar R, Lim JS, Yap TA . Towards Precision Medicine in the Clinic: From Biomarker Discovery to Novel Therapeutics. Trends Pharmacol Sci 2017; 38: 25–40.
Sholl LM, Do K, Shivdasani P, Cerami E, Dubuc AM, Kuo FC et al. Institutional implementation of clinical tumor profiling on an unselected cancer population. JCI Insight 2016; 1: e87062.
Gerlinger M, Rowan AJ, Horswell S, Larkin J, Endesfelder D, Gronroos E et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med 2012; 366: 883–892.
Miller CA, Gindin Y, Lu C, Griffith OL, Griffith M, Shen D et al. Aromatase inhibition remodels the clonal architecture of estrogen-receptor-positive breast cancers. Nature Communications 2016; 7: 12498.
Yates LR, Gerstung M, Knappskog S, Desmedt C, Gundem G, Van Loo P et al. Subclonal diversification of primary breast cancer revealed by multiregion sequencing. Nat Med 2015; 21: 751–759.
Kostadinov R, Maley CC, Kuhner MK . Bulk Genotyping of Biopsies Can Create Spurious Evidence for Hetereogeneity in Mutation Content. PLoS Comput Biol 2016; 12: e1004413.
Brastianos PK, Carter SL, Santagata S, Cahill DP, Taylor-Weiner A, Jones RT et al. Genomic Characterization of Brain Metastases Reveals Branched Evolution and Potential Therapeutic Targets. Cancer Discovery 2015; 5: 1164–1177.
Harper KL, Sosa MS, Entenberg D, Hosseini H, Cheung JF, Nobre R et al. Mechanism of early dissemination and metastasis in Her2+ mammary cancer. Nature 2016 doi:10.1038/nature20609.
Hosseini H, Obradovic MM, Hoffmann M, Harper KL, Sosa MS, Werner-Klein M et al. Early dissemination seeds metastasis in breast cancer. Nature 2016 doi:10.1038/nature20785.
McDonald OG, Li X, Saunders T, Tryggvadottir R, Mentch SJ, Warmoes MO et al. Epigenomic reprogramming during pancreatic cancer progression links anabolic glucose metabolism to distant metastasis. Nat Genet 2017; 49: 367–376.
Liu X, Ory V, Chapman S, Yuan H, Albanese C, Kallakury B et al. ROCK inhibitor and feeder cells induce the conditional reprogramming of epithelial cells. Am J Pathol 2012; 180: 599–607.
Brodt P . Role of the Microenvironment in Liver Metastasis: From Pre- to Prometastatic Niches. Clin Cancer Res 2016; 22: 5971–5982.
Nakshatri H, Anjanappa M, Bhat-Nakshatri P . Ethnicity-Dependent and -Independent Heterogeneity in Healthy Normal Breast Hierarchy Impacts Tumor Characterization. Scientific Reports 2015; 5: 13526.
Cancer Genome Atlas Research N. Comprehensive molecular characterization of gastric adenocarcinoma. Nature 2014; 513: 202–209.
Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discovery 2012; 2: 401–404.
Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Science Signaling 2013; 6: pl1.
Pepe C, Guidugli L, Sensi E, Aretini P, D'Andrea E, Montagna M et al. Methyl group metabolism gene polymorphisms as modifier of breast cancer risk in Italian BRCA1/2 carriers. Breast Cancer Res Treat 2007; 103: 29–36.
Flowers M, Birkey Reffey S, Mertz SA . Marc Hurlbert for the Metastatic Breast Cancer A. Obstacles, Opportunities and Priorities for Advancing Metastatic Breast. Cancer Research. Cancer Res 2017; 77: 3386–3390.
Visvader JE, Stingl J . Mammary stem cells and the differentiation hierarchy: current status and perspectives. Genes Dev 2014; 28: 1143–1158.
Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF . Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci U S A 2003; 100: 3983–3988.
Sheridan C, Kishimoto H, Fuchs RK, Mehrotra S, Bhat-Nakshatri P, Turner CH et al. CD44+/CD24- breast cancer cells exhibit enhanced invasive properties: an early step necessary for metastasis. Breast Cancer Res 2006; 8: R59.
Kim J, Villadsen R, Sorlie T, Fogh L, Gronlund SZ, Fridriksdottir AJ et al. Tumor initiating but differentiated luminal-like breast cancer cells are highly invasive in the absence of basal-like activity. Proc Natl Acad Sci U S A 2012; 109: 6124–6129.
Cancer Genome Atlas N. Comprehensive molecular portraits of human breast tumours. Nature 2012; 490: 61–70.
Sebestyen E, Zawisza M, Eyras E . Detection of recurrent alternative splicing switches in tumor samples reveals novel signatures of cancer. Nucleic Acids Res 2015; 43: 1345–1356.
Stephens PJ, Tarpey PS, Davies H, Van Loo P, Greenman C, Wedge DC et al. The landscape of cancer genes and mutational processes in breast cancer. Nature 2012; 486: 400–404.
Fujimoto A, Totoki Y, Abe T, Boroevich KA, Hosoda F, Nguyen HH et al. Whole-genome sequencing of liver cancers identifies etiological influences on mutation patterns and recurrent mutations in chromatin regulators. Nat Genet 2012; 44: 760–764.
Chmielecki J, Crago AM, Rosenberg M, O'Connor R, Walker SR, Ambrogio L et al. Whole-exome sequencing identifies a recurrent NAB2-STAT6 fusion in solitary fibrous tumors. Nat Genet 2013; 45: 131–132.
Robinson DR, Wu YM, Kalyana-Sundaram S, Cao X, Lonigro RJ, Sung YS et al. Identification of recurrent NAB2-STAT6 gene fusions in solitary fibrous tumor by integrative sequencing. Nat Genet 2013; 45: 180–185.
Gruber TA, Downing JR . The biology of pediatric acute megakaryoblastic leukemia. Blood 2015; 126: 943–949.
Kumar R, Manning J, Spendlove HE, Kremmidiotis G, McKirdy R, Lee J et al. ZNF652, a novel zinc finger protein, interacts with the putative breast tumor suppressor CBFA2 T3 to repress transcription. Mol Cancer Res 2006; 4: 655–665.
van Bon BW, Oortveld MA, Nijtmans LG, Fenckova M, Nijhof B, Besseling J et al. CEP89 is required for mitochondrial metabolism and neuronal function in man and fly. Human Molecular Genetics 2013; 22: 3138–3151.
Vaque JP, Dorsam RT, Feng X, Iglesias-Bartolome R, Forsthoefel DJ, Chen Q et al. A genome-wide RNAi screen reveals a Trio-regulated Rho GTPase circuitry transducing mitogenic signals initiated by G protein-coupled receptors. Mol Cell 2013; 49: 94–108.
Chen D, Huang X, Cai J, Guo S, Qian W, Wery JP et al. A set of defined oncogenic mutation alleles seems to better predict the response to cetuximab in CRC patient-derived xenograft than KRAS 12/13 mutations. Oncotarget 2015; 6: 40815–40821.
Park JT, Johnson N, Liu S, Levesque M, Wang YJ, Ho H et al. Differential in vivo tumorigenicity of diverse KRAS mutations in vertebrate pancreas: A comprehensive survey. Oncogene 2015; 34: 2801–2806.
Odore E, Lokiec F, Cvitkovic E, Bekradda M, Herait P, Bourdel F et al. Phase I Population Pharmacokinetic Assessment of the Oral Bromodomain Inhibitor OTX015 in Patients with Haematologic Malignancies. Clin Pharmacokinet 2016; 55: 397–405.
Bafna S, Kaur S, Batra SK . Membrane-bound mucins: the mechanistic basis for alterations in the growth and survival of cancer cells. Oncogene 2010; 29: 2893–2904.
Mitchell RA, Metz CN, Peng T, Bucala R . Sustained mitogen-activated protein kinase (MAPK) and cytoplasmic phospholipase A2 activation by macrophage migration inhibitory factor (MIF). Regulatory role in cell proliferation and glucocorticoid action. J Biol Chem 1999; 274: 18100–18106.
Hu X, Lu H, Cao S, Deng YL, Li QJ, Wan Q et al. Stem cells derived from human first-trimester umbilical cord have the potential to differentiate into oocyte-like cells in vitro. Int J Mol Med 2015; 35: 1219–1229.
Petit FM, Serres C, Bourgeon F, Pineau C, Auer J . Identification of sperm head proteins involved in zona pellucida binding. Hum Reprod 2013; 28: 852–865.
Cormier S, Vandormael-Pournin S, Babinet C, Cohen-Tannoudji M . Developmental expression of the Notch signaling pathway genes during mouse preimplantation development. Gene Expr Patterns 2004; 4: 713–717.
Giampieri R, Scartozzi M, Loretelli C, Piva F, Mandolesi A, Lezoche G et al. Cancer stem cell gene profile as predictor of relapse in high risk stage II and stage III, radically resected colon cancer patients. PloS One 2013; 8: e72843.
Steeg PS, Camphausen KA, Smith QR . Brain metastases as preventive and therapeutic targets. Nat Rev Cancer 11: 352–363.
Burnett RM, Craven KE, Krishnamurthy P, Goswami CP, Badve S, Crooks P et al. Organ-specific adaptive signaling pathway activation in metastatic breast cancer cells. Oncotarget 2015; 6: 12682–12696.
Chen L, Liu P, Evans Jr TC, Ettwiller LM . DNA damage is a pervasive cause of sequencing errors, directly confounding variant identification. Science 2017; 355: 752–756.
Coomans de Brachene A, Demoulin JB . FOXO transcription factors in cancer development and therapy. Cellular and Molecular Life Sciences: CMLS 2016; 73: 1159–1172.
Pereira B, Chin SF, Rueda OM, Vollan HK, Provenzano E, Bardwell HA et al. The somatic mutation profiles of 2433 breast cancers refines their genomic and transcriptomic landscapes. Nature Communications 2016; 7: 11479.
Vainchenker W, Kralovics R . Genetic basis and molecular pathophysiology of classical myeloproliferative neoplasms. Blood 2017; 129: 667–679.
Eirew P, Steif A, Khattra J, Ha G, Yap D, Farahani H et al. Dynamics of genomic clones in breast cancer patient xenografts at single-cell resolution. Nature 2015; 518: 422–426.
Cheung LW, Yu S, Zhang D, Li J, Ng PK, Panupinthu N et al. Naturally occurring neomorphic PIK3R1 mutations activate the MAPK pathway, dictating therapeutic response to MAPK pathway inhibitors. Cancer Cell 2014; 26: 479–494.
McCreery MQ, Halliwill KD, Chin D, Delrosario R, Hirst G, Vuong P et al. Evolution of metastasis revealed by mutational landscapes of chemically induced skin cancers. Nat Med 2015; 21: 1514–1520.
Cejalvo JM, Martinez de Duenas E, Galvan P, Garcia-Recio S, Burgues Gasion O, Pare L et al. Intrinsic Subtypes and Gene Expression Profiles in Primary and Metastatic Breast Cancer. Cancer Res 2017; 77: 2213–2221.
Kishi N, Tang Z, Maeda Y, Hirai A, Mo R, Ito M et al. Murine homologs of deltex define a novel gene family involved in vertebrate Notch signaling and neurogenesis. Int J Dev Neurosci 2001; 19: 21–35.
Linch SN, Kasiewicz MJ, McNamara MJ, Hilgart-Martiszus IF, Farhad M, Redmond WL . Combination OX40 agonism/CTLA-4 blockade with HER2 vaccination reverses T-cell anergy and promotes survival in tumor-bearing mice. Proc Natl Acad Sci U S A 2016; 113: E319–E327.
Arun B, Kilic G, Yen C, Foster B, Yardley DA, Gaynor R et al. Loss of FHIT expression in breast cancer is correlated with poor prognostic markers. Cancer Epidemiol Biomarkers Prev 2005; 14: 1681–1685.
Sinha R, Hussain S, Mehrotra R, Kumar RS, Kumar K, Pande P et al. Kras gene mutation and RASSF1A, FHIT and MGMT gene promoter hypermethylation: indicators of tumor staging and metastasis in adenocarcinomatous sporadic colorectal cancer in Indian population. PloS One 2013; 8: e60142.
Yan W, Xu N, Han X, Zhou XM, He B . The clinicopathological significance of FHIT hypermethylation in non-small cell lung cancer, a meta-analysis and literature review. Scientific Reports 2016; 6: 19303.
Buxton IL, Yokdang N, Matz RM . Purinergic mechanisms in breast cancer support intravasation, extravasation and angiogenesis. Cancer Lett 2010; 291: 131–141.
Jozwik KM, Chernukhin I, Serandour AA, Nagarajan S, Carroll JS . FOXA1 Directs H3K4 Monomethylation at Enhancers via Recruitment of the Methyltransferase MLL3. Cell Reports 2016; 17: 2715–2723.
Zhang Z, Christin JR, Wang C, Ge K, Oktay MH, Guo W . Mammary-Stem-Cell-Based Somatic Mouse Models Reveal Breast Cancer Drivers Causing Cell Fate Dysregulation. Cell Reports 2016; 16: 3146–3156.
Cheng J, Blum R, Bowman C, Hu D, Shilatifard A, Shen S et al. A role for H3K4 monomethylation in gene repression and partitioning of chromatin readers. Mol Cell 2014; 53: 979–992.
Kloosterman WP, Hoogstraat M, Paling O, Tavakoli-Yaraki M, Renkens I, Vermaat JS et al. Chromothripsis is a common mechanism driving genomic rearrangements in primary and metastatic colorectal cancer. Genome Biol 2011; 12: R103.
Hao Y, Zhang P, Xuei X, Nakshatri H, Edenberg HJ, Li L et al. Statistical modeling for sensitive detection of low-frequency single nucleotide variants. BMC Genomics 2016; 17 (Suppl 7): 514.
Hao YXX, Li L, Nakshatri H, Edenberg HJ, Liu Y . A framework for detecting low frequency single nucleotide variants. Journal of Computational Biology 2017; 24: 637–646.
Fisher MM, Watkins RA, Blum J, Evans-Molina C, Chalasani N, DiMeglio LA et al. Elevations in Circulating Methylated and Unmethylated Preproinsulin DNA in New-Onset Type 1 Diabetes. Diabetes 2015; 64: 3867–3872.
Acknowledgements
We thank tissue collection team at the IU Simon Cancer Center, Clinical Research Office and Neurooncology Center at IU School of Medicine for collection of fresh tissues for the study. We also thank the flow cytometry core at the IU Simon Cancer Center. Excellent support from New York Genome Center, particularly Mr Benjamin Hubert, is highly appreciated. IUPUI Signature Center for the Cure of Glioblastoma supported brain metastases tissue collection. Susan G Komen for the Cure (SAC110025 to HN), Indiana CTSI Project development pilot grant (to HN, LL and KNP) and IU Simon Cancer Center Breast Cancer Program Pilot grant (to YL and HN) supported this study. This study utilized core services by National Institutes of Health Grant P30 DK097512 to the Indiana University School of Medicine.
Author contributions
MA, primary cell culturing, DNA and RNA extraction and flow cytometry; YH, RareVar development, bioinformatics and analyses of genomic data; ERS, bioinformatics and analyses of genomic data; PN, primary cell collection and DNA/RNA extraction; JN, digital droplet PCR; SAT, digital droplet PCR, RGM, digital droplet PCR assay design and implementation; AAC, patient accrual, collection and processing of brain metastases; MRS, clinical protocol development and brain metastases collection; LL, bioinformatics and analyses of genomic data; FF, pathway analyses of genomic data; KPN, pathway analyses of genomic data and manuscript writing; KDM, clinical protocol development and implementation of liver metastasis sample collection; YL, assay design, supervision of bioinformatics efforts and manuscript writing; HN, experimental design, primary cell culturing, data interpretation, manuscript writing and overall supervision of the project. All authors read and approve the manuscript.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Oncogene website
Supplementary information
Rights and permissions
About this article
Cite this article
Anjanappa, M., Hao, Y., Simpson, E. et al. A system for detecting high impact-low frequency mutations in primary tumors and metastases. Oncogene 37, 185–196 (2018). https://doi.org/10.1038/onc.2017.322
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/onc.2017.322
This article is cited by
-
Nonsynonymous, synonymous and nonsense mutations in human cancer-related genes undergo stronger purifying selections than expectation
BMC Cancer (2019)
-
Attraction and Compaction of Migratory Breast Cancer Cells by Bone Matrix Proteins through Tumor-Osteocyte Interactions
Scientific Reports (2018)
-
Patient-derived conditionally reprogrammed cells maintain intra-tumor genetic heterogeneity
Scientific Reports (2018)
-
Organotropism: new insights into molecular mechanisms of breast cancer metastasis
npj Precision Oncology (2018)
-
Targeting epigenetics using synthetic lethality in precision medicine
Cellular and Molecular Life Sciences (2018)