Abstract
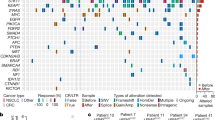
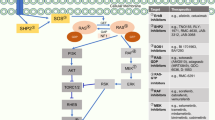
There are currently no effective targeted therapies for KRAS mutant cancers. Therapeutic strategies that combine MEK inhibitors with agents that target apoptotic pathways may be a promising therapeutic approach. We investigated combining MEK and MDM2 inhibitors as a potential treatment strategy for KRAS mutant non-small cell lung cancers (NSCLC) and colorectal carcinomas that harbor wild-type TP53. The combination of pimasertib (MEK inhibitor) and SAR405838 (MDM2 inhibitor) was synergistic and induced the expression of PUMA and BIM, led to apoptosis and growth inhibition in vitro, and tumor regression in vivo. Acquired resistance to the combination commonly resulted from the acquisition of TP53 mutations, conferring complete resistance to MDM2 inhibition. In contrast, resistant clones exhibited marked variability in sensitivity to MEK inhibition, which significantly impacted sensitivity to subsequent treatment with alternative MEK inhibitor-based combination therapies. These results highlight both the potential promise and limitations of combining MEK and MDM2 inhibitors for treatment of KRAS mutant NSCLC and colorectal cancers.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Kobayashi S, Boggon TJ, Dayaram T, Janne PA, Kocher O, Meyerson M et al. EGFR mutation and resistance of non-small-cell lung cancer to gefitinib. N Engl J Med 2005; 352: 786–792.
Maemondo M, Inoue A, Kobayashi K, Sugawara S, Oizumi S, Isobe H et al. Gefitinib or chemotherapy for non-small-cell lung cancer with mutated EGFR. N Engl J Med 2010; 362: 2380–2388.
Flaherty KT, Robert C, Hersey P, Nathan P, Garbe C, Milhem M et al. Improved survival with MEK inhibition in BRAF-mutated melanoma. N Engl J Med 2012; 367: 107–114.
Chapman PB, Hauschild A, Robert C, Haanen JB, Ascierto P, Larkin J et al. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N Engl J Med 2011; 364: 2507–2516.
Prior IA, Lewis PD, Mattos C . A comprehensive survey of Ras mutations in cancer. Cancer Res 2012; 72: 2457–2467.
Blumenschein GR Jr., Smit EF, Planchard D, Kim DW, Cadranel J, De Pas T et al. A randomized phase II study of the MEK1/MEK2 inhibitor trametinib (GSK1120212) compared with docetaxel in KRAS-mutant advanced non-small-cell lung cancer (NSCLC)dagger. Ann Oncol 2015; 26: 894–901.
Engelman JA, Chen L, Tan X, Crosby K, Guimaraes AR, Upadhyay R et al. Effective use of PI3K and MEK inhibitors to treat mutant Kras G12D and PIK3CA H1047R murine lung cancers. Nat Med 2008; 14: 1351–1356.
Corcoran RB, Cheng KA, Hata AN, Faber AC, Ebi H, Coffee EM et al. Synthetic lethal interaction of combined BCL-XL and MEK inhibition promotes tumor regressions in KRAS mutant cancer models. Cancer Cell 2013; 23: 121–128.
Sun C, Hobor S, Bertotti A, Zecchin D, Huang S, Galimi F et al. Intrinsic resistance to MEK inhibition in KRAS mutant lung and colon cancer through transcriptional induction of ERBB3. Cell Rep 2014; 7: 86–93.
Brown CJ, Lain S, Verma CS, Fersht AR, Lane DP . Awakening guardian angels: drugging the p53 pathway. Nat Rev Cancer 2009; 9: 862–873.
Hainaut P, Hollstein M . p53 and human cancer: the first ten thousand mutations. Adv Cancer Res 2000; 77: 81–137.
Muller PA, Vousden KH . p53 mutations in cancer. Nat Cell Biol 2013; 15: 2–8.
Wu X, Bayle JH, Olson D, Levine AJ . The p53-mdm-2 autoregulatory feedback loop. Genes Dev 1993; 7: 1126–1132.
Bond GL, Hu W, Levine AJ . MDM2 is a central node in the p53 pathway: 12 years and counting. Curr Cancer Drug Targets 2005; 5: 3–8.
Momand J, Wu HH, Dasgupta G . MDM2–master regulator of the p53 tumor suppressor protein. Gene 2000; 242: 15–29.
Oliner JD, Kinzler KW, Meltzer PS, George DL, Vogelstein B . Amplification of a gene encoding a p53-associated protein in human sarcomas. Nature 1992; 358: 80–83.
Ding L, Getz G, Wheeler DA, Mardis ER, McLellan MD, Cibulskis K et al. Somatic mutations affect key pathways in lung adenocarcinoma. Nature 2008; 455: 1069–1075.
Stott FJ, Bates S, James MC, McConnell BB, Starborg M, Brookes S et al. The alternative product from the human CDKN2A locus, p14(ARF), participates in a regulatory feedback loop with p53 and MDM2. EMBO J 1998; 17: 5001–5014.
Shangary S, Qin D, McEachern D, Liu M, Miller RS, Qiu S et al. Temporal activation of p53 by a specific MDM2 inhibitor is selectively toxic to tumors and leads to complete tumor growth inhibition. Proc Natl Acad Sci USA 2008; 105: 3933–3938.
Shangary S, Wang S . Small-molecule inhibitors of the MDM2-p53 protein-protein interaction to reactivate p53 function: a novel approach for cancer therapy. Annu Rev Pharmacol Toxicol 2009; 49: 223–241.
Tovar C, Graves B, Packman K, Filipovic Z, Higgins B, Xia M et al. MDM2 small-molecule antagonist RG7112 activates p53 signaling and regresses human tumors in preclinical cancer models. Cancer Res 2013; 73: 2587–2597.
Vassilev LT . MDM2 inhibitors for cancer therapy. Trends Mol Med 2007; 13: 23–31.
Vassilev LT, Vu BT, Graves B, Carvajal D, Podlaski F, Filipovic Z et al. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science 2004; 303: 844–848.
Wang S, Sun W, Zhao Y, McEachern D, Meaux I, Barriere C et al. SAR405838: an optimized inhibitor of MDM2-p53 interaction that induces complete and durable tumor regression. Cancer Res 2014; 74: 5855–5865.
Ji Z, Njauw CN, Taylor M, Neel V, Flaherty KT, Tsao H . p53 rescue through HDM2 antagonism suppresses melanoma growth and potentiates MEK inhibition. J Invest Dermatol 2012; 132: 356–364.
Ji Z, Kumar R, Taylor M, Rajadurai A, Marzuka-Alcala A, Chen YE et al. Vemurafenib synergizes with nutlin-3 to deplete survivin and suppresses melanoma viability and tumor growth. Clin Cancer Res 2013; 19: 4383–4391.
Saiki AY, Caenepeel S, Yu D, Lofgren JA, Osgood T, Robertson R et al. MDM2 antagonists synergize broadly and robustly with compounds targeting fundamental oncogenic signaling pathways. Oncotarget 2014; 5: 2030–2043.
Hata AN, Yeo A, Faber AC, Lifshits E, Chen Z, Cheng KA et al. Failure to induce apoptosis via BCL-2 family proteins underlies lack of efficacy of combined MEK and PI3K inhibitors for KRAS-mutant lung cancers. Cancer Res 2014; 74: 3146–3156.
Vousden KH, Lane DP . p53 in health and disease. Nat Rev Mol Cell Biol 2007; 8: 275–283.
Caunt CJ, Sale MJ, Smith PD, Cook SJ . MEK1 and MEK2 inhibitors and cancer therapy: the long and winding road. Nat Rev Cancer 2015; 15: 577–592.
Munoz-Espin D, Serrano M . Cellular senescence: from physiology to pathology. Nat Rev Mol Cell Biol 2014; 15: 482–496.
Villunger A, Michalak EM, Coultas L, Mullauer F, Bock G, Ausserlechner MJ et al. p53- and drug-induced apoptotic responses mediated by BH3-only proteins puma and noxa. Science 2003; 302: 1036–1038.
Miyashita T, Reed JC . Tumor suppressor p53 is a direct transcriptional activator of the human bax gene. Cell 1995; 80: 293–299.
Aziz MH, Shen H, Maki CG . Acquisition of p53 mutations in response to the non-genotoxic p53 activator Nutlin-3. Oncogene 2011; 30: 4678–4686.
Michaelis M, Rothweiler F, Barth S, Cinatl J, van Rikxoort M, Loschmann N et al. Adaptation of cancer cells from different entities to the MDM2 inhibitor nutlin-3 results in the emergence of p53-mutated multi-drug-resistant cancer cells. Cell Death Dis 2011; 2: e243.
Ebi H, Corcoran RB, Singh A, Chen Z, Song Y, Lifshits E et al. Receptor tyrosine kinases exert dominant control over PI3K signaling in human KRAS mutant colorectal cancers. J Clin Invest 2011; 121: 4311–4321.
Kitai H, Ebi H, Tomida S, Floros KV, Kotani H, Adachi Y et al. Epithelial-to-Mesenchymal transition defines feedback activation of receptor tyrosine kinase signaling induced by MEK inhibition in KRAS-mutant lung cancer. Cancer Discov 2016; 6: 754–769.
Turke AB, Song Y, Costa C, Cook R, Arteaga CL, Asara JM et al. MEK inhibition leads to PI3K/AKT activation by relieving a negative feedback on ERBB receptors. Cancer Res 2012; 72: 3228–3237.
Ray-Coquard I, Blay JY, Italiano A, Le Cesne A, Penel N, Zhi J et al. Effect of the MDM2 antagonist RG7112 on the P53 pathway in patients with MDM2-amplified, well-differentiated or dedifferentiated liposarcoma: an exploratory proof-of-mechanism study. Lancet Oncol 2012; 13: 1133–1140.
Zhang W, Konopleva M, Burks JK, Dywer KC, Schober WD, Yang JY et al. Blockade of mitogen-activated protein kinase/extracellular signal-regulated kinase kinase and murine double minute synergistically induces Apoptosis in acute myeloid leukemia via BH3-only proteins Puma and Bim. Cancer Res 2010; 70: 2424–2434.
Wade M, Li YC, Wahl GM . MDM2, MDMX and p53 in oncogenesis and cancer therapy. Nat Rev Cancer 2013; 13: 83–96.
Hoffman-Luca CG, Yang CY, Lu J, Ziazadeh D, McEachern D, Debussche L et al. Significant differences in the development of acquired resistance to the MDM2 inhibitor SAR405838 between in vitro and in vivo drug treatment. PLoS ONE 2015; 10: e0128807.
Jung J, Lee JS, Dickson MA, Schwartz GK, Le Cesne A, Varga A et al. TP53 mutations emerge with HDM2 inhibitor SAR405838 treatment in de-differentiated liposarcoma. Nat Commun 2016; 7: 12609.
Manchado E, Weissmueller S, Morris JPt, Chen CC, Wullenkord R, Lujambio A et al. A combinatorial strategy for treating KRAS-mutant lung cancer. Nature 2016; 534: 647–651.
Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S et alSTAR: ultrafast universal RNA-seq alignerBioinformatics 2013; 29: 15–21.
Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc 2012; 7: 562–578.
Liao Y, Smyth GK, Shi W . featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014; 30: 923–930.
Robinson MD, McCarthy DJ, Smyth GK . edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010; 26: 139–140.
Ritz C, Streibig J . From additivity to synergism – a modelling perspective. Synergy 2014; 1: 22–29.
R_Core_Team R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing: Vienna, Austria, 2015.
Bolger AM, Lohse M, Usadel B . Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 2014; 30: 2114–2120.
Mose LE, Wilkerson MD, Hayes DN, Perou CM, Parker JS . ABRA: improved coding indel detection via assembly-based realignment. Bioinformatics 2014; 30: 2813–2815.
Cibulskis K, Lawrence MS, Carter SL, Sivachenko A, Jaffe D, Sougnez C et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol 2013; 31: 213–219.
Wilm A, Aw PP, Bertrand D, Yeo GH, Ong SH, Wong CH et al. LoFreq: a sequence-quality aware, ultra-sensitive variant caller for uncovering cell-population heterogeneity from high-throughput sequencing datasets. Nucleic acids Res 2012; 40: 11189–11201.
Ye K, Schulz MH, Long Q, Apweiler R, Ning Z . Pindel: a pattern growth approach to detect break points of large deletions and medium sized insertions from paired-end short reads. Bioinformatics 2009; 25: 2865–2871.
Ramos AH, Lichtenstein L, Gupta M, Lawrence MS, Pugh TJ, Saksena G et al. Oncotator: cancer variant annotation tool. Hum Mutat 2015; 36: E2423–E2429.
Acknowledgements
We thank all members of the Engelman Lab for helpful discussions and feedback. We thank MercK KGaA, Darmstadt, Germany for provision of pimasertib for our research and for scientific review of this manuscript. This study was funded by support from the NIH R01CA14059408 (JAE), Uniting Against Lung Cancer (ANH), Conquer Cancer Foundation of ASCO (ANH).
Author contributions
ANH, JW and JAE designed the study, analyzed the data and wrote the paper. ANH, FMS, HLA and MGC performed cell line and biochemical studies. MGC performed cell line tumor xenograft studies. SR performed drug synergy analysis. JJ, SR, FJ and RIS performed RNA-sequencing and analysis. ML, JSL and JJ performed TP53 genotyping of cell lines. LD and SS generated the CRC PDX model and performed in vivo studies. All authors discussed the results and commented on the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
JAE is a consultant for Novartis, Sanofi, Genentech and Amgen; has research agreements with Novartis, Sanofi, and Amgen. ANH has provided consulting services for Amgen; has research agreements with Amgen and Novartis. SR, ML, JSL, JJ, SS, LD, and JW are employees of Sanofi, as noted in the affiliations. No other conflicts of interest were reported by the other authors.
Additional information
Supplementary Information accompanies this paper on the Oncogene website
Supplementary information
Rights and permissions
About this article
Cite this article
Hata, A., Rowley, S., Archibald, H. et al. Synergistic activity and heterogeneous acquired resistance of combined MDM2 and MEK inhibition in KRAS mutant cancers. Oncogene 36, 6581–6591 (2017). https://doi.org/10.1038/onc.2017.258
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/onc.2017.258
This article is cited by
-
Combined MEK/MDM2 inhibition demonstrates antitumor efficacy in TP53 wild-type thyroid and colorectal cancers with MAPK alterations
Scientific Reports (2022)
-
Resistance mechanisms to inhibitors of p53-MDM2 interactions in cancer therapy: can we overcome them?
Cellular & Molecular Biology Letters (2021)
-
Treatment scheduling effects on the evolution of drug resistance in heterogeneous cancer cell populations
npj Breast Cancer (2021)
-
β2-AR blockade potentiates MEK1/2 inhibitor effect on HNSCC by regulating the Nrf2-mediated defense mechanism
Cell Death & Disease (2020)
-
Tetra-substituted pyrrole derivatives act as potent activators of p53 in melanoma cells
Investigational New Drugs (2020)