Abstract
Isoprenylcysteine carboxylmethyltransferase (ICMT) catalyzes the post-translational modification of RAB GTPases that contain C-terminal CXC motifs. However, the functional impact of this modification on RAB proteins has not been actively explored. We found that inhibition of ICMT significantly reduced cell migration in vitro and cancer invasion and metastasis in vivo. This role of ICMT was found to be mediated by RAB4A, an essential regulator of the fast recycling of integrin β3. Integrin β3 regulates cell polarity and migration when localized appropriately to the plasma membrane, thereby having an essential role in cancer metastasis. ICMT catalyzed carboxylmethylation is critical for RAB4A activation and interaction with effectors, its localization to endosomes and recycling vesicles, and hence important for RAB4A-dependent integrin β3 recycling to plasma membrane. These findings bring attention to the effects of C-terminal carboxylmethylation on RAB GTPases and provide a rationale for targeting ICMT in the treatment of metastatic cancer.
Similar content being viewed by others
Introduction
Isoprenylcysteine carboxylmethyltransferase (ICMT) catalyzes the final step of post-translational prenylation processing of a select group of proteins that contain C-terminal CAAX or CXC consensus motifs.1 This modification and its functional impact on CAAX proteins such as RAS have been extensively studied,2, 3, 4 mostly focusing on cancer cell proliferation and survival.3, 5, 6, 7, 8, 9 In comparison, there has been no in depth study on the cellular consequences of carboxylmethylation of CXC substrates and their roles in cancer progression,1 although it has been biochemically demonstrated that these GTPases undergo modification by ICMT.10
RAB4A, a CXC RAB protein, is first geranylgeranylated by RAB geranylgeranyltransferase on both cysteines, then carboxylmethylated on the C-terminal cysteine by ICMT.2, 10, 11, 12 As a GTPase, the activity of RAB4A depends on the guanosine triphosphate/guanosine diphosphate (GTP/GDP)-binding state, which is affected by multiple regulators such as GTPase-activating proteins, guanine nucleotide exchange factors and GDP dissociation inhibitors (GDI).13 RAB4A regulates intracellular vesicular trafficking, particularly the recycling from early endosome to plasma membrane.
Metastasis is the main cause of cancer mortality.14 The process of tumor cell invasion into surrounding tissue, intravasation into and extravasation out of vasculature, and establishing secondary colonies constitute key steps in cancer progression in which the interaction between cell surface integrins and extracellular matrix has important roles.15, 16, 17 Among different subtypes, integrin β3 is found to have consistently high expression in a variety of aggressive cancers.18, 19, 20, 21, 22 Integrin recycling, a dynamic process that selectively removes integrin from one part of the plasma membrane by endocytosis followed by its re-deposition to another, is critical for integrin activation in migrating cell.23 Studies have shown that RAB4A is indispensable for the growth factor-stimulated ‘short loop’ recycling of integrin β3 from early endosome to the leading edge of cell to support directional movement.24, 25 The potential importance of RAB4A in the process of invasion and metastasis in cancer cells is supported by the findings that its expression increases progressively from normal tissue to primary tumor to metastatic tumor.26
In this study, we sought to investigate the role of ICMT-mediated methylation on RAB4A function in promoting the dynamic recycling of integrin β3, and further on the biological process of cancer cell metastasis.
Results
ICMT function is important for cancer cell migration and metastasis
Given the suggested role of ICMT in cell movement,27 we examined the impact of ICMT inhibition on cancer cell metastasis. Time-lapse migration analysis of MDA-MB-231, a highly metastatic human breast cancer cell line, revealed that the speed and linear persistence of migration were reduced significantly when ICMT expression is suppressed (Figures 1a–c). Transwell migration assays also demonstrated that ICMT suppression, either by short hairpin RNA (shRNA) knockdown or by ICMT inhibitor treatment, attenuated MDA-MB-231 cell migration (Figure 1d). Similar reduction of migration was observed in HT-1080 fibrosarcoma cells when ICMT was suppressed (Supplementary Figure S1). The morphology of cell clusters in three-dimensional matrigel culture can be used as a read-out for their metastatic potential.28 HT-1080 cells grown in matrigel formed cell clusters with a complex branching/stellate multi-cellular structure commonly associated with invasiveness, whereas cells expressing ICMT shRNA formed small, round cell aggregates (Figure 1e).
Icmt inhibition suppresses cancer cell migration in vitro and metastasis in vivo. (a, b) Time-lapse imaging of MDA-MB-231 cell movement by IncuCyte ZOOM (Essen Bioscience). Attached cells expressing either control or Icmt shRNA were imaged for 24 h at 30-min intervals. Thirty cells were tracked in each population and analyzed by ImageJ (http://rsbweb.nih.gov/ij/) to calculate distances of migration (b). Results are expressed as mean±s.d., n=30 cells for each group; ***P<0.001. (c) Quantitative real-time RT–PCR analysis of Icmt knockdown efficiency for the cells in a and b. Results are expressed as mean±s.d.; n=3 for each group, ***P<0.001. (d) Transwell migration study of MDA-MB-231 cells. The comparisons are made between the control cells and those in which Icmt function has been inhibited, either by shRNA targeting Icmt or by cysmethynil treatment as indicated. Data shown are from one experiment that has been repeated three times with similar results. (e) HT-1080 fibrosarcoma cell cluster morphology. Cells expressing either control or Icmt shRNA were grown in three-dimensional matrigel and imaged after 1 and 6 days. Images shown are from one experiment that has been repeated with similar results. (f) Chicken CAM metastasis model. Human tumor cells were grafted onto the CAM at embryonic day 10 and allowed to form a tumor by incubating the inoculated eggs for an additional up to 7 days. Upper CAM and lower CAM were imaged by dissecting microscope to identify invasion of cancer cells. (g, h) In vivo invasion and metastasis of GFP-expressing HT-1080 cells in CAM assay. Cells expressing either control or Icmt shRNA were seeded on the CAM of developing chick embryos and visualized in the upper CAM (g) and lower CAM (h) after 5 and 7 days, respectively. Each image is representative of five such fields sampled per embryo, out of nine embryos in each group. (i) Lung metastasis of luciferase-expressing HT-1080 cells from xenografts. HT-1080-Luc cells (1 × 106), expressing either control or Icmt shRNA, were injected in the flank of five SCID mice each. The animals were killed after primary tumors reached the size of 15 × 15 mm3 and ex vivo images of lungs are presented. N=5 mice each were used for HT-1080-Luc cells expressing control shRNA and those expressing shRNA targeting Icmt. No randomization is necessary for this study.
To investigate the impact of ICMT in vivo, we made use of two metastasis models. The first is the chick chorioallantoic membrane (CAM) assay, which is used to study invasion of colonized human cancer cells into the proximal and distal sites of chorioallantoic membrane of fertilized eggs (Figure 1f). Briefly, CAM (shown as the red lining in the Figure 1f) is the vascularized extra-embryonic membrane connected to the embryo via the allantoic vein. CAM is therefore an accessible and visible metastasis target site for experimental manipulation and observations. Metastasized cancer cells on the membrane can be found near the primary tumor (upper CAM) and farther away from it (lower CAM). The fertilized eggs are first incubated for 10 days, followed by seeding of cancer cells near the junction of blood vessels. The endpoint analysis for the metastasis of HT-1080 cells to upper CAM is on day 5 and to lower CAM on day 7 after seeding.29 In the protocol used here, HT-1080 cells expressing green fluorescent protein (GFP) and either control shRNA or shRNA targeting Icmt were seeded onto the upper CAM ex ovo near the allantoic vein on embryonic day 10. Five days after tumor cell seeding, visible primary tumors were formed for both cell populations (Figure 1g). For the control tumor, significant numbers of cancer cells invaded into the surrounding membrane (upper CAM) and metastasized to the distal membrane (lower CAM) at days 5 and 7, respectively, whereas those expressing Icmt shRNA were largely contained within the primary tumor (Figures 1g and h).
In vivo metastasis was also investigated in SCID mouse xenograft model using HT-1080 cells. Ex vivo lung bioluminescence imaging analysis for luciferase-expressing HT-1080 cells detected substantially higher numbers of metastasis foci in the lungs of mice harboring tumors from cells expressing control shRNA (Figure 1i, top row) than from those expressing shRNA targeting Icmt (Figure 1i, bottom row). Thus, both in vitro and in vivo studies support the critical role of ICMT function in human cancer cell metastasis.
Integrin β3 promotion of cell migration is ICMT dependent
Integrin β3 has been consistently linked to invasiveness and cancer mortality of many different tissue types.19, 20, 21, 22 Activated plasma membrane integrin clusters and signals inwardly leading to focal adhesion kinase (FAK) activation, establishment of cell polarity and promotion of cell movement.30, 31 In both HT-1080 and MDA-MB-231 cells, inhibition of Icmt, either by knockdown or inhibitor treatment, resulted in significant reduction of plasma membrane localization of integrin β3 and FAK activation, and disruption of normal actin cytoskeletal organization with clear loss of stress fibers and cell polarity (Figure 2a, HT-1080 cells; Supplementary Figure S2, MDA-MB-231 cells). Clustering of integrin β3 in response to Mn2+ treatment, which stimulates the activation and aggregation of integrin β3 molecules localized on the plasma membrane,32 was also abolished with the suppression of ICMT function (Figures 2b and c). Hence, results from both approaches showed that ICMT function is essential for the plasma membrane localization and activation of integrin β3.
Integrin β3-driven cancer cell invasiveness is ICMT dependent. (a) Analysis of cellular localization of integrin β3, p-FAK and F-actin in HT-1080 cells. Cells expressing either control or Icmt shRNA (left), or treated by either vehicle or cysmethynil (right) were stained with anti-β3 integrin (BD Biosciences), anti-p-FAK or Texas Red-conjugated phalloidin for actin as indicated, and 4’, 6-diamidino-2-phenylindole (DAPI) for nucleus before fluorescence confocal microscopy imaging. (b, c) Fluorescent microscopy analysis of β3 integrin clustering stimulated by Mn2+ in HT-1080 cells. The effect of ICMT suppression on β3 integrin clustering was observed in cells expressing either control or Icmt shRNA (b), or treated with either vehicle or cysmethynil for 24 h (c), before MnCl2 (0.5 mM) stimulation for 30 min. The cells were stained with anti-β3 integrin antibody (Abcam) and DAPI. (d) Immunoblot analysis of integrin β3 protein levels in MCF-7 and MDA-MB-231 cells (left), and in MCF-7 cells without or with exogenous expression of integrin β3 (right). pITGB3: vector containing integrin β3 coding cDNA; pEV: empty vector. (e) Cell morphology of MCF-7 without or with exogenous integrin β3 expression. (f) Fluorescent microscopy localization analysis of integrin β3, p-FAK and F-actin in parental MCF-7 cells or those overexpressing integrin β3, after 24-h treatment by either vehicle control or cysmethynil. Antibodies for integrin β3, p-FAK or F-actin are the same as a. (g) Top: fluorescent microscopy analysis of MCF-7 cells with or without exogenous expression of integrin β3 and with or without the expression of Icmt shRNA. Integrin β3 antibody is the same as in a. Bottom: transwell migration analysis was performed on the cells as the top panels. (h) Quantification analysis for the transwell study in g after crystal violet staining the lower membrane of the transwell chamber and visualization at 540 nm. Results are expressed as mean±s.d. of three wells for each condition; ***P<0.001. Experiments have been repeated three times with similar results.
To investigate the functional impact of ICMT on integrin β3 regulation of cell migration and invasion, we studied the breast cancer cell line MCF-7, which expresses much lower levels of integrin β3 (Figure 2d, left) and has limited metastatic potential compared with MDA-MB-231 cells. Enforced expression of integrin β3 in MCF-7 cells (Figure 2d, right) resulted in a remarkable change of MCF-7 cells to a more polarized morphology (Figure 2e), increased levels of p-FAK and actin stress fiber formation (Figure 2f). Importantly, all these changes were abolished by treatment with ICMT inhibitor (Figure 2f). Similar to inhibitor treatment, shRNA knockdown of Icmt obliterated cell polarity (Figure 2g) and enhanced directional migration resulted from higher level of integrin β3 expression, as measured by transwell migration assay (Figures 2g and h). Together, the studies with MCF-7 cells demonstrated that integrin β3 function in promoting cancer cell polarity and migration is ICMT dependent.
RAB4A, a substrate of ICMT, has an essential role in cancer cell migration and metastasis by facilitating integrin β3 membrane localization
RAB4A is involved in the dynamic recycling of integrin β3 from early endosome to plasma membrane in response to pro-migratory stimuli such as platelet-derived growth factor (PDGF), which promotes ‘short loop’ recycling of integrin β3 and cell migration.24, 25, 33 In MDA-MB-231 and HT-1080 cells, knockdown of Icmt strongly attenuated the leading edge localization of integrin β3 and RAB4 both at basal state and under PDGF stimulation (Figure 3a, MDA-MB-231 cells; Supplementary Figure S3A, HT-1080 cells), suggesting an essential role for ICMT in the function of RAB4A regulation of integrin β3 localization. In contrast, RAB5—a key regulator for endocytosis34 that is not a substrate of ICMT—remained colocalized with early endosome marker EEA1 with or without Icmt knockdown (Supplementary Figure S3B).
ICMT regulation of plasma membrane localization of integrin β3 is mediated by RAB4, which function to promote cancer cell metastasis. (a) Immunofluorescent analysis of endogenous integrin β3 and RAB4 localization in MDA-MB-231 cells expressing either control or Icmt shRNA targeting, with or without 30 min PDGF stimulation, before prepared for confocal microscopy analysis. (b) Immunofluorescent analysis for integrin β3 localization and F-actin organization in HT-1080 cells transfected with either control siRNA or RAB4A siRNA. Antibody for integrin β3 and Texas Alexa Fluor 488 phalloidin are the same as in Figure 2. (c) CAM in vivo metastasis study on GFP-expressing HT-1080 cells with or without RAB4A knockdown. The experiment was conducted as illustrated in Figure 1f. Each image is representative of at least six from one embryo; there are five embryos for each group.
We further studied whether the role in regulating integrin β3 recycling is specific for RAB4A, one of the two isoforms of RAB4. RNA interference knockdown of RAB4A abolished integrin β3 recruitment at leading edge, cell polarity and disrupted actin cytoskeleton organization in HT-1080 cells (Figure 3b). These phenotypes were confirmed with two sequence distinct targeting small interfering RNAs (siRNAs) in MDA-MB-231 cells (Supplementary Figure S4A). In contrast, knockdown of RAB4B did not have the same impact (Supplementary Figure S4B), consistent with the notion that RAB4A, but not RAB4B, regulates integrin β3 localization and function. In agreement with reduction of integrin β3 signaling, the levels p-FAK and p-paxillin were reduced in the cells transfected with RAB4A siRNA (Supplementary Figure S4C).
The role of RAB4A in migration and invasion was validated using the CAM in vivo metastasis model. GFP-expressing HT-1080 cells were transfected with either control siRNA or that targeting RAB4A before being seeded for CAM study on embryonic day 10. Primary tumors were formed 5 days after grafting for both cell populations (Figure 3c). In the control siRNA group, many cells had disseminated from the primary tumor to the surrounding membrane (upper CAM) and to distal sites (lower CAM) 5 days after seeding, respectively, whereas the cells in the RAB4A siRNA group were mostly contained within the primary tumor (Figure 3c). Thus, the CAM study demonstrated that, consistent with the in vitro findings, RAB4A is important for cancer cell metastasis in vivo.
Modification by ICMT is important for RAB4A function in the regulation of integrin β3 plasma membrane localization and cell migration
To assess the role of RAB4A in the observed ICMT regulation of metastasis, we investigated the ability of RAB4A to rescue the directional migration of HT-1080 cells suppressed by Icmt knockdown. The enforced expression of RAB4A significantly rescued the migration defect (Figures 4a–c). At the cellular level, RAB4A overexpression also restored the plasma membrane localization of integrin β3, the organization of actin stress fibers and cell polarity in cells in which Icmt expression was suppressed (Figure 4d). As ICMT functions to methylate C-terminus cysteine of RAB4A, we examined the ability of mutant RAB4A lacking the C-terminal CXC (RAB4A-ΔCXC) to rescue the phenotypes in Icmt knockdown cells. In contrast to the rescue seen with wild-type RAB4A (Figures 4a, b and d), expression of RAB4A-ΔCXC rescued neither the defect of directional migration, nor the integrin β3 localization and actin organization phenotypes arising from Icmt silencing (Figures 4e–h). On the contrary, RAB4A-ΔCXC appeared to have some dominant-negative effect on directional migration of cells. Taken together, the rescue studies demonstrate that the post-translational modification of the RAB4A CXC motif is critical for RAB4A function in the regulation of integrin β3 localization and cancer cell migration.
Wild-type, not CXC deleted RAB4A, restores integrin β3 plasma membrane localization and cancer cell migration suppressed because of ICMT deficiency. (a, b) Ectopic expression of RAB4A rescues the migration defect resulted from Icmt knockdown in HT-1080 cells. Icmt knockdown was done using Icmt shRNA. Wild-type RAB4A complementary DNA was introduced in retrovirus. Quantification of cell migration in matrigel was as described earlier. Data were expressed as mean±s.d. of four determinations from each group; ***P<0.001. (c) Immunoblot analysis of RAB4 protein levels in control and RAB4A OE cells from a and b. (d) Localization of integrin β3, RAB4 and F-actin in the four groups of HT-1080 cells prepared as in a. Immunofluorescent confocal microscopy analysis was performed as in Figure 2. (e–h) Data shown here are from experiments performed the same way as in a–d, respectively. However, instead of wild-type RAB4A, Rab4A-ΔCXC, which is lacking C-terminal 3 amino acids, was expressed. All experiments were repeated three times; the images are representative from at least 30 such images taken from the three experiments.
ICMT promotes integrin β3 recycling by regulating RAB4A activation
Based on the evidence thus far, it seemed likely that ICMT regulates integrin β3 recycling. Indeed, using an enzyme-linked immunosorbent assay capture assay,24 Icmt knockdown was found to reduce integrin β3 recycling from intracellular compartment to cell surface (Figure 5a). Analysis of the levels of GTP/GDP-bound RAB4A revealed that the levels of activated RAB4A were reduced significantly in Icmt knockdown cells compared with that of control cells, both at basal state and following stimulation with PDGF (Figure 5b), suggesting that C-terminal carboxylmethylation is important for the activation of RAB4A.
ICMT impacts integrin β3 recycling to plasma membrane by regulating RAB4A activation, binding to interacting proteins, and early endosome recycling. (a) Integrin β3 recycling in HT-1080 cells expressing either control shRNA or that targeting Icmt. The recycling assay was performed as previously reported.24 Results are expressed as mean±s.d. of three technical repeats for each group; **P<0.01. (b) Quantification of active RAB4A in HT-1080 cells with or without Icmt knockdown under basal cultural condition or with PDGF stimulation as described for Figure 3b. RAB4A activation assay is as described in Materials and methods section. (c, d) Co-immunoprecipitation of exogenously expressed (c) or endogenous (d) RAB4A and RABAPTIN-5 in HT-1080 cells. Cell lysates were prepared for immunoprecipitation with anti-Flag (c) or anti-active RAB4A antibody (d), followed by immunoblot analysis with anti-Rabaptin-5 antibody. Quantification of the protein band intensity was done using ImageJ software; the ratios of intensities of pulldown vs total Rabaptin-5 are presented. (e) PLA of the interaction of endogenous RAB4A and RABAPTIN-5 in HT-1080 cells infected with either control shRNA or shRNA targeting Icmt. Images are representative of at least 10 fields per condition; the experiment was repeated twice with similar results. (f, g) Co-immunoprecipitation of RAB4 and RAB-GDI in HT-1080 cells. Pulldown was performed using either anti-GDI (f) or anti-RAB4 (g) antibody, followed by immunoblot analysis with anti-RAB4 (f) or anti-RAB-GDI (g) antibody, respectively. Protein band intensities and ratios were determined as in c and d. (h) PLA of the interaction of endogenous RAB4A and RAB-GDI in HT-1080 cells. (i) Visualization of recycling vesicles in HT-1080 cells. Retrovirus expressing wild-type RAB4A, dominant-negative mutant RAB4A-S27N, or the empty vector control (EV) was introduced into HT-1080 cells stably expressing either control or Icmt shRNA. The cells were processed for immunofluorescent confocal microscopy for active RAB4A (red) and AP1 (green). (j) Analysis for integrin β3 localization in HT-1080 cells by immunofluorescence confocal microscopy; anti-β3 integrin (red) and DAPI (blue). For all microscopy analysis, images shown are representatives of least 50 cells counted in at least 10 fields for each condition. The experiments were repeated twice with similar findings.
RABAPTIN-5 is a direct downstream effector of activated RAB4A in vesicle recycling.35, 36 Consistent with the notion that ICMT is important for RAB4A activation, co-immunoprecipitation analysis demonstrated that significantly less RABAPTIN-5 was associated with RAB4A in Icmt knockdown cells compared with control cells (Figures 5c and d). Proximity ligation assay (PLA), a method to study close proximity of two proteins, also suggested that the interaction between RABAPTIN-5 and RAB4A was significantly reduced in Icmt knockdown cells (Figure 5e). Finally, immunofluorescence analysis showed significant level of colocalization between RAB4A and RABAPTIN-5 in control cells in what are presumed to be recycling vesicles, and that this colocalization was markedly reduced in Icmt knockdown cells (Supplementary Figure S5). The RAB regulator GDI preferentially interacts with GDP-bound RAB proteins.37, 38, 39 Indeed, there was markedly higher level of co-immunoprecipitation of RAB4 and GDI in cells with Icmt knockdown compared with control cells (Figures 5f and g). The PLA study consistently showed significant elevation of RAB4A and RAB-GDI interaction in Icmt knockdown cells (Figure 5h). Taken together, the interaction studies support the role of ICMT modification in RAB4A activation and its interaction with downstream effectors and regulators.
RAB4A functions to promote short loop recycling of select cargo proteins such as integrin β3. Adaptor-related protein complex 1 (AP1) is recruited to recycling endosomes as an essential component in vesicle transport.40, 41 An increase in vesicles positive for both AP1 and RAB4A suggests an elevated state of fast recycling from endosome to the plasma membrane. Knockdown of Icmt reduced the number of cell cortical vesicles that are positive for AP1 and RAB4A (Figure 5i). Exogenous expression of wild-type RAB4A, but not the inactive RAB4A-S27N, markedly increased the number of AP1- and RAB4A-positive vesicles at the periphery of the cells (Figure 5i), suggesting a compensatory state of stimulated recycling. Consistent with the notion that activation is necessary for its function, RAB4A-S27N, unlike wild-type RAB4A, failed to restore integrin β3 plasma membrane localization suppressed by Icmt knockdown (Figure 5j). Notably, neither RAB5A nor RAB11A, which are not substrates for ICMT modification,12 could restore the phenotypes of recycling vesicle formation, leading edge integrin β3 localization and cell polarity suppressed by ICMT inhibition (Supplementary Figure S6), which is compelling evidence that ICMT regulates β3 integrin fast recycling by enhancing the function of RAB4A, a CXC substrate of ICMT.
Integrin β3 and its recycling are relevant predictors for breast cancer patient survival
Data presented so far have made a strong case for the importance of ICMT and its substrate RAB4A in cancer metastasis. We further investigated the relevance of integrin β3 recycling in the survival of cancer patients. A previous study reported that RAB4A expression is amplified in various tumors.26 Building on this, we queried the cBio Cancer Genomics Portal for gene alterations of RAB4A and integrin β3 in invasive breast cancers. The analysis revealed that either RAB4A or integrin β3 is amplified, and more so when both are accounted for, in a variety of tumors, especially in invasive breast cancers (Figure 6a–c). Analysis of the breast cancer database using Geneanalytics tool (http://geneanalytics.duhs.duke.edu) revealed that high expression of RAB4A or AP1G1 (AP1 gamma 1 subunit, a core component of the AP1 complex) correlates with poorer prognosis (Figures 6d and e). As RAB4A and AP1 are important in the formation and function of recycling vesicles, transporting integrin to the plasma membrane, the survival data suggest that the process of recycling is relevant for the breast cancer progression and patient survival.
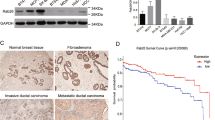
RAB4A and integrin β3 are amplified in invasive breast cancers, and RAB4A and AP1G1 expression predicts poor clinical outcome in breast cancer patients. (a–c) RAB4A (a), integrin β3 (b) and either RAB4A or integrin β3 (c) gene changes are analyzed in various cancers, particularly invasive breast cancers. The cBio Cancer Genomics Portal (http://www.cbioportal.org) was queried for RAB4A, integrin β3, and both RAB4A and integrin β3 for genetic changes. The tumor data sets were collected from the Cancer Genome Atlas (TCGA), the US National Cancer Institute (NCI) and others that reported in the bio Cancer Genomics Portal. (d, e) Kaplan–Meier plots for 609 breast cancer patients stratified by either RAB4A or AP1G1 expression using the breast cancer data sets that were collected and analyzed in the Geneanalytics tool (http://geneanalytics.duhs.duke.edu). The survival data were grouped according to ‘high’ and ‘low’ expression categories using the median splits. Y axes = the probability of relapse free survival. X axes = months of follow up. P-values are from a log-rank test, N=609.
Discussion
The development of therapies against cancer metastasis has lagged behind that against proliferation. Anti-proliferative drugs have generally not shown beneficial effects on metastasis in preclinical models.42, 43 The development of targeted therapy for the molecules that are essential for the process of metastasis presents as a challenging opportunity. Integrins, for example, have been targeted as novel metastasis therapeutics in clinical trials.44, 45 However, it appears that inhibiting integrin trafficking may be more effective than direct targeting.46
Previous studies demonstrated that C-terminal CXC motif of RAB4 protein is double geranylgeranylated and carboxylmethylated on the terminal cysteine.12, 47 Geranylgeranylation of the CXC motif has been shown to be functionally important for RAB4A,48 however, the functional consequences of carboxylmethylation by ICMT have not been investigated in cancer. This study reveals that ICMT function is essential in the recycling and plasma membrane localization of integrin β3, focal adhesion assembly and organization of actin cytoskeleton, hence for cell polarity and migration.
RAB4A activity is regulated by the GTP/GDP cycle and GDI binding. Our data indicate ICMT regulates RAB4A activation, as there is a reduction of the activated form of RAB4A and increased interaction between RAB4A and GDI when ICMT is inhibited. The reduction of activated RAB4A on the endosomal membrane negatively impacts the early endosomal sorting machinery through RAB4A/RABAPTIN-5/AP1 signaling, resulting in deficient β3 integrin recycling to plasma membrane. This study not only extends the understanding of biological impact of ICMT beyond that of CAAX proteins, but also makes a case for ICMT as a potential target for novel therapies against the process of metastasis.
Materials and methods
Cell culture and treatment conditions
The human breast cancer cell lines MDA-MB-231, MCF-7, human fibrosarcoma cell line HT-1080 and human embryonic kidney HEK293T cells were obtained from the American Type Culture Collection (Rockville, MD, USA). Cells were cultured in the Dulbecco’s modified Eagle's medium (Gibco, Gaithersburg, MD, USA) with fetal bovine serum (Hyclone, Logan, UT, USA), penicillin and streptomycin (Gibco) following ATCC standard conditions. Cysmethynil, a small molecule inhibitor of ICMT, was synthesized in the Duke Chemical Biology Synthesis Lab (Duke University, Durham, NC, USA) and dissolved in dimethyl sulfoxide. The final treatment concentrations were made in culture medium containing 5% serum. Cells were subjected to treatment for 24–48 h before subsequent analysis, except for the experiments described in Figure 1d where the cells were pre-treated with 17.5 μM cysmethynil for 24 h before transwell migration assay. The concentrations of cysmethynil used in the study have been shown to inhibit ICMT carboxylmethylation in cells in a target-specific manner.49 To assay Mn2+-induced β3 integrin activation and clustering, the cells were stimulated with 0.5 mM Mn2+ for 30 min after respective pretreatment in each experiment. To study PDGF-induced β3 integrin plasma membrane localization, the serum-starved cells were first stimulated with 20 ng/ml PDGF-BB for 30 min followed by immunofluorescent assay for integrin β3.
Quantitative real-time RT–PCR analysis and siRNAs
RNA and complementary DNA were prepared from cells using RNeasy mini kit (Qiagen, Hilden, Germany) and Maxima H minus First Strand cDNA Synthesis kit (Thermo Fisher Scientific, Waltham, MA, USA) according to the manufacturer’s protocol. Quantitative PCR was performed using SYBR green (Thermo Fisher Scientific) with the following primers: human Icmt: forward, 5′-GTTTCGGCATCCTTCTTACG-3′; reverse, 5′-CACTGTCAGGGCATAGCTGA-3′; 18S ribosomal RNA: forward, 5′-AAGTTCGACCGTCTTCTCAGC-3′; and reverse, 5′-GTTGATTAAGTCCCTGCCCTTTG-3′. RAB4A siRNA sequences are: 5′-CACCGUUAGAUGUGUAUG-3′ and 5′-UUACAUACACAUCUAACG-3′.
Immunoprecipitation and western blot analysis
Cell lysates were prepared, and protein concentrations determined at 562 nm using the BCA protein assay kit. Protein A agarose beads (25 μl) were pre-incubated with 1 μl of anti-rabbit Rab4 antibody (Abcam, Cambridge, UK) for 3 h at 4 °C on rotator, whereupon concentration adjusted lysate was added to the mixture and incubated overnight at 4 °C. Each sample of immunoprecipitant was collected and washed three times with lysis buffer, resuspended in sodium dodecyl sulfate sample buffer and heated for 5 min at 95 °C before analysis by sodium dodecyl sulfate–polyacrylamide gel electrophoresis and immunoblot with the specified antibodies.8 Anti-β3 integrin (ab75872) and anti-Rab4 (ab13252) were from Abcam; anti-phospho FAK Tyr397 (MAB1144) was from Millipore (Billerica, MA, USA); anti-phospho paxillin Ser178 (A300-100A) was from Bethyl Laboratories (Montgomery, TX, USA); anti-paxillin (2542) and anti-GAPDH (2118) were from Cell Signalling Technologies (Beverly, MA, USA); anti-Rab-GDI 1 (ABIN1742290) was from Synaptic System (GmbH, Gottingen, Germany); anti-Rabaptin-5 (610676) was from BD Biosciences, San Jose, CA, USA.
Transfection and establishing stable cell lines
Transfection was performed using Lipofectamin 2000 transfection reagent (Invitrogen, Carlsbad, CA, USA) by standard lab protocols.9 To create stable HT-1080 cells expressing firefly luciferase, parental cells were transfected with pMSCV-Luciferase vector and single cell clones were selected by Geneticin (Life Technologies, Carlsbad, CA, USA), and expression level was quantified using IVIS 200 imaging system (PerkinElmer, Waltham, MA, USA). To generate MCF-7 cells expressing GFP-integrin β3, the parental cells were first transfected with the construct50 as described, followed by selection in Geneticin (Life Technologies) containing medium. To generate ICMT knockdown cell line, lentiviruses were first produced by transfection of HEK293T cells with pRSV-Rev, pMD2.G-VSV.g, pMDLg/pRRE (Addgene, Cambride, MA, USA) and lentiviral vector pLL3.7 containing the shRNA targeting human ICMT sequence 5′-CTTGGTTTCGGCATCCTTCTT-3′, per procedure described previously.51 The lentiviruses thus produced were used to infect HT-1080 or MDA-MB-231.
To generate RAB4A-overexpressing cell lines, we cloned wild-type Rab4A, Rab4A-ΔCXC and Rab4A-S27N coding sequences into retroviral vector pMSCV between XhoI and EcoRI sites. HEK293T cells were then transfected with pCL-Ampho and above pMSCV plasmids to produce viruses that were used to infect HT-1080 and MDA-MB231 cells using standard polybrene protocol. Rab4A-ΔCXC and Rab4A-S27N mutant sequences were generated using Site-Directed Mutagenesis Kit following the manufacturer’s instructions (Agilent Technologies, Santa Clara, CA, USA).
Immunofluorescent and confocal microscopy
Immunofluorescent and confocal microscopies were performed by established protocols.52 The antibodies include: anti-β3 integrin (555752), anti-EEA1 (610456) and anti-Rabaptin-5 (610676) are from BD Biosciences; anti-β3 integrin (ab75872); anti-Rab4 (ab13252), anti-Rab5 (ab13253) and anti-gamma Adaptin (ab21980) are from Abcam; anti-Rab4A (21067) and anti-active Rab4A (26921) are from NewEast Biosciences, Malvern, PA, USA; anti-phospho FAK Tyr397 (MAB1144) is from Millipore. F-actin was detected using Alexa Fluor 488 phalloidin (A12379) and Alexa Fluor 568 phalloidin (A12380) from Thermo Fisher Scientific. Samples were analyzed using LSM710 Carl Zeiss Confocal Microscope (Olympus, Shinjuku, Tokyo, Japan) equipped with an Olympus 60 x oil immersion objective.
Cell migration assays
Transwell migration assay was carried out using the transwell chamber with polycarbonate membrane having pore size of 8.0 μm (Corning Life Sciences, Oneonta, NY, USA). The bottom chamber was filled with Dulbecco’s modified Eagle's medium containing 20% fetal bovine serum and the upper chamber was seeded with 5 × 104 cells in standard Dulbecco’s modified Eagle's medium. The experiments were performed according to the user manual of Corning Life Sciences. After 48-h incubation and removal of the cells on top of the membrane, the cells on the bottom side of the membrane were fixed and stained with crystal violet, followed by light microscope imaging. Absorption at optical density 540 nm was also measured for quantification.
Cell migration was also analyzed by wound assay, which was measured by IncuCyte ZOOM live cell imaging system (Essen Bioscience, Ann Arbor, MI, USA). HT-1080 cells or MDA-MB-231 cells were seeded in the vitronectin-coated (Sigma, Steinheim, Germany) 96-well ImageLock plate (Essen Bioscience). Cells were allowed to grow in standard tissue culture condition overnight, followed by scratching on cell monolayer using WoundMaker (Essen Bioscience). After two phosphate-buffered saline washes and addition of fresh medium, the plate was placed into IncuCyte for timed imaging every 30 min for 48 h.
Three-dimensional matrigel assay
HT-1080 cells were seeded in matrigel (BD Biosciences) coated 96-well plate (50 μl per well). After 20-min incubation, the standard medium was replaced by that containing 5% Matrigel. Cells were cultured and fluorescence microscopy images were obtained (IX71S1F3, Olympus) on days 1 and 6.
Integrin β3 internalization and recycling assay
Integrin β3 internalization and recycling assay was done following a previously described surface biotinylation and capture–enzyme-linked immunosorbent assay method.24 MesNa, iodoacetamide and detection buffer are from Sigma; Maxisorb 96-well plates for capture–enzyme-linked immunosorbent assay are from Life Technologies; anti-integrin β3 antibody is from BD Biosciences; streptavidin-conjugated horseradish peroxidase is from Amersham (Biosciences, Bucks, UK).
Proximity ligation assay
PLA is a method to study whether two proteins are in close proximity by detecting amplified fluorescent signals from a modified PCR reaction. We followed the instruction manual of Duolink kit (Sigma-Aldrich, Steinheim, Germany). Briefly, cells were seeded on eight-well chamber slide (Thermo Fisher Scientific), cultured for 24 h, fixed with 4% formaldehyde and permeabilized with 0.2% Triton X-100 in phosphate-buffered saline for 5 min, followed by blocking and incubation with primary antibody pairs of rabbit anti-Rab4A (NewEast Biosciences) with either mouse Rab-GDI (Synaptic System), or mouse anti-Rabaptin-5 (BD Biosciences). The corresponding DuoLink In Situ PLA probes (containing oligonucleotides) were then added 1 h before mixing with ligase; the reaction was allowed to proceed to amplify the Alexa Fluor 568 signal for 100 min. The signals were visualization by fluorescent microscopy.
RAB4A activation assay
RAB4A activation status was assessed by RAB4A-GTP pulldown using RAB4A Activation Kit according to the manufacturer’s protocol (NewEast Biosciences). Briefly, the cell lysate was mixed with anti-Rab4A-GTP monoclonal antibody pre-incubated with protein A/G Agarose beads, and the mixture was incubated overnight at 4 °C with gentle agitation. The beads were pelleted by centrifugation for 1 min at 5000 × g, and washed three times with 0.5 ml of assay buffer. The collected pellet was processed for standard sodium dodecyl sulfate–polyacrylamide gel electrophoresis and immunoblot procedures for the detection of Rab4A protein.
CAM metastasis model
We have followed the published method.29 Briefly, fertilized eggs were incubated in a rotating incubator (Brinsea, Titusville, FL, USA) at 37 °C with 70% humidity. The CAM was lowered from the shell and a small window was created over the region near allantoic vein at embryonic day 10. One million HT-1080 cells stably expressing GFP were seeded on the CAM after a gentle swipe by a sterile cotton swab on the CAM. The windows over the area were then taped over and the eggs were returned to the incubator. The embryos were killed 5 or 7 days after implantation of the tumor cells; the GFP-positive metastasized tumor cells in upper and lower CAM were visualized using Stereozoom dissecting microscope (SZX16, Olympus).
SCID mouse spontaneous metastasis study
Luciferase-expressing HT-1080 cells were infected with lentiviruses expressing either control shRNA or that targeting Icmt. Five days post-infection, 1 × 106 cells in 100 μl of serum-free Dulbecco’s modified Eagle's medium containing 30% matrigel were deposited subcutaneously into the flank of 8-week-old female SCID mouse. In this study, endpoint ex vivo lung metastasis was quantified by image analysis of the number and intensity of luminescence foci. Each primary tumor, either from cells expressing control shRNA or those expressing shRNA targeting Icmt, was allowed to develop to the size of 15 × 15 mm3, whereupon the mouse was killed 15 min after intraperitoneal injection of 200 μl of 15 mg/ml d-luciferin solution (PerkinElmer). The killed animal was subjected to immediate post-mortal surgery to remove lungs for ex vivo bioluminescence imaging using the IVIS Lumina system (Xenogen, PerkinElmer). Animals were maintained by procedures approved by our Institutional Animal Care and Use Committee (IACUC) for the humane treatment of laboratory animals.
Statistical analysis
All statistical analysis in this study was performed using GraphPad InStat (GraphPad Software, CA, USA). Data are presented as mean±s.d. Relapse-free survival curves were calculated according to the Kaplan–Meier method, and comparison was performed using the log-rank test. A two-tailed unpaired Student’s t-test was used for other statistical analyses. Statistical significance was defined as *P<0.05, **P<0.01, ***P<0.001.
References
Wang M, Casey PJ . Protein prenylation: unique fats make their mark on biology. Nat Rev Mol Cell Biol 2016; 17: 110–122.
Bergo MO, Leung GK, Ambroziak P, Otto JC, Casey PJ, Young SG . Targeted inactivation of the isoprenylcysteine carboxyl methyltransferase gene causes mislocalization of K-Ras in mammalian cells. J Biol Chem 2000; 275: 17605–17610.
Cushman I, Casey PJ . Role of isoprenylcysteine carboxylmethyltransferase-catalyzed methylation in Rho function and migration. J Biol Chem 2009; 284: 27964–27973.
Hanker AB, Mitin N, Wilder RS, Henske EP, Tamanoi F, Cox AD et al. Differential requirement of CAAX-mediated posttranslational processing for Rheb localization and signaling. Oncogene 2010; 29: 380–391.
Bergo MO, Gavino BJ, Hong C, Beigneux AP, McMahon M, Casey PJ et al. Inactivation of Icmt inhibits transformation by oncogenic K-Ras and B-Raf. J Clin Invest 2004; 113: 539–550.
Lau HY, Ramanujulu PM, Guo D, Yang T, Wirawan M, Casey PJ et al. An improved isoprenylcysteine carboxylmethyltransferase inhibitor induces cancer cell death and attenuates tumor growth in vivo. Cancer Biol Ther 2014; 15: 1280–1291.
Teh JT, Zhu WL, Ilkayeva OR, Li Y, Gooding J, Casey PJ et al. Isoprenylcysteine carboxylmethyltransferase regulates mitochondrial respiration and cancer cell metabolism. Oncogene 2015; 34: 3296–3304.
Wang M, Tan W, Zhou J, Leow J, Go M, Lee HS et al. A small molecule inhibitor of isoprenylcysteine carboxymethyltransferase induces autophagic cell death in PC3 prostate cancer cells. J Biol Chem 2008; 283: 18678–18684.
Wang M, Hossain MS, Tan W, Coolman B, Zhou J, Liu S et al. Inhibition of isoprenylcysteine carboxylmethyltransferase induces autophagic-dependent apoptosis and impairs tumor growth. Oncogene 2010; 29: 4959–4970.
Smeland TE, Seabra MC, Goldstein JL, Brown MS . Geranylgeranylated Rab proteins terminating in Cys-Ala-Cys, but not Cys-Cys, are carboxyl-methylated by bovine brain membranes in vitro. Proc Natl Acad Sci USA 1994; 91: 10712–10716.
Bergo MO, Leung GK, Ambroziak P, Otto JC, Casey PJ, Gomes AQ et al. Isoprenylcysteine carboxyl methyltransferase deficiency in mice. J Biol Chem 2001; 276: 5841–5845.
Leung KF, Baron R, Ali BR, Magee AI, Seabra MC . Rab GTPases containing a CAAX motif are processed post-geranylgeranylation by proteolysis and methylation. J Biol Chem 2007; 282: 1487–1497.
Stenmark H . Rab GTPases as coordinators of vesicle traffic. Nat Rev Mol Cell Biol 2009; 10: 513–525.
Weigelt B, Peterse JL, van 't Veer LJ . Breast cancer metastasis: markers and models. Nat Rev Cancer 2005; 5: 591–602.
Desgrosellier JS, Cheresh DA . Integrins in cancer: biological implications and therapeutic opportunities. Nat Rev Cancer 2010; 10: 9–22.
Guo W, Giancotti FG . Integrin signalling during tumour progression. Nat Rev Mol Cell Biol 2004; 5: 816–826.
Webb DJ, Parsons JT, Horwitz AF . Adhesion assembly, disassembly and turnover in migrating cells — over and over and over again. Nat Cell Biol 2002; 4: E97–100.
Brooks PC, Clark RA, Cheresh DA . Requirement of vascular integrin alpha v beta 3 for angiogenesis. Science 1994; 264: 569–571.
Gruber G, Hess J, Stiefel C, Aebersold DM, Zimmer Y, Greiner RH et al. Correlation between the tumoral expression of beta3-integrin and outcome in cervical cancer patients who had undergone radiotherapy. Br J Cancer 2005; 92: 41–46.
Hosotani R, Kawaguchi M, Masui T, Koshiba T, Ida J, Fujimoto K et al. Expression of integrin alphaVbeta3 in pancreatic carcinoma: relation to MMP-2 activation and lymph node metastasis. Pancreas 2002; 25: e30–e35.
McCabe NP, De S, Vasanji A, Brainard J, Byzova TV . Prostate cancer specific integrin alphavbeta3 modulates bone metastatic growth and tissue remodeling. Oncogene 2007; 26: 6238–6243.
Sloan EK, Pouliot N, Stanley KL, Chia J, Moseley JM, Hards DK et al. Tumor-specific expression of alphavbeta3 integrin promotes spontaneous metastasis of breast cancer to bone. Breast Cancer Res 2006; 8: R20.
Paul NR, Jacquemet G, Caswell PT . Endocytic trafficking of integrins in cell migration. Curr Biol 2015; 25: R1092–R1105.
Roberts M, Barry S, Woods A, van der Sluijs P, Norman J . PDGF-regulated rab4-dependent recycling of alphavbeta3 integrin from early endosomes is necessary for cell adhesion and spreading. Curr Biol 2001; 11: 1392–1402.
White DP, Caswell PT, Norman JC . Alpha v beta3 and alpha5beta1 integrin recycling pathways dictate downstream Rho kinase signaling to regulate persistent cell migration. J Cell Biol 2007; 177: 515–525.
Frittoli E, Palamidessi A, Marighetti P, Confalonieri S, Bianchi F, Malinverno C et al. A RAB5/RAB4 recycling circuitry induces a proteolytic invasive program and promotes tumor dissemination. J Cell Biol 2014; 206: 307–328.
Cushman I, Casey PJ . RHO methylation matters: a role for isoprenylcysteine carboxylmethyltransferase in cell migration and adhesion. Cell Adh Migr 2011; 5: 11–15.
Lee GY, Kenny PA, Lee EH, Bissell MJ . Three-dimensional culture models of normal and malignant breast epithelial cells. Nat Methods 2007; 4: 359–365.
Zijlstra A, Mellor R, Panzarella G, Aimes RT, Hooper JD, Marchenko ND et al. A quantitative analysis of rate-limiting steps in the metastatic cascade using human-specific real-time polymerase chain reaction. Cancer Res 2002; 62: 7083–7092.
Frame MC, Patel H, Serrels B, Lietha D, Eck MJ . The FERM domain: organizing the structure and function of FAK. Nat Rev Mol Cell Biol 2010; 11: 802–814.
Serrels B, Serrels A, Brunton VG, Holt M, McLean GW, Gray CH et al. Focal adhesion kinase controls actin assembly via a FERM-mediated interaction with the Arp2/3 complex. Nat Cell Biol 2007; 9: 1046–1056.
Legler DF, Wiedle G, Ross FP, Imhof BA . Superactivation of integrin alphavbeta3 by low antagonist concentrations. J Cell Sci 2001; 114: 1545–1553.
Woods AJ, White DP, Caswell PT, Norman JC . PKD1/PKCmu promotes alphavbeta3 integrin recycling and delivery to nascent focal adhesions. EMBO J 2004; 23: 2531–2543.
Bucci C, Parton RG, Mather IH, Stunnenberg H, Simons K, Hoflack B et al. The small GTPase rab5 functions as a regulatory factor in the early endocytic pathway. Cell 1992; 70: 715–728.
Deneka M, Neeft M, Popa I, van Oort M, Sprong H, Oorschot V et al. Rabaptin-5alpha/rabaptin-4 serves as a linker between rab4 and gamma(1)-adaptin in membrane recycling from endosomes. EMBO J 2003; 22: 2645–2657.
Vitale G, Rybin V, Christoforidis S, Thornqvist P, McCaffrey M, Stenmark H et al. Distinct Rab-binding domains mediate the interaction of Rabaptin-5 with GTP-bound Rab4 and Rab5. EMBO J 1998; 17: 1941–1951.
Araki S, Kikuchi A, Hata Y, Isomura M, Takai Y . Regulation of reversible binding of smg p25A, a ras p21-like GTP-binding protein, to synaptic plasma membranes and vesicles by its specific regulatory protein, GDP dissociation inhibitor. J Biol Chem 1990; 265: 13007–13015.
Ignatev A, Kravchenko S, Rak A, Goody RS, Pylypenko O . A structural model of the GDP dissociation inhibitor rab membrane extraction mechanism. J Biol Chem 2008; 283: 18377–18384.
Pfeffer S, Aivazian D . Targeting Rab GTPases to distinct membrane compartments. Nat Rev Mol Cell Biol 2004; 5: 886–896.
D'Souza RS, Semus R, Billings EA, Meyer CB, Conger K, Casanova JE . Rab4 orchestrates a small GTPase cascade for recruitment of adaptor proteins to early endosomes. Curr Biol 2014; 24: 1187–1198.
Folsch H, Ohno H, Bonifacino JS, Mellman I . A novel clathrin adaptor complex mediates basolateral targeting in polarized epithelial cells. Cell 1999; 99: 189–198.
Ratajczak MZ, Jadczyk T, Schneider G, Kakar SS, Kucia M . Induction of a tumor-metastasis-receptive microenvironment as an unwanted and underestimated side effect of treatment by chemotherapy or radiotherapy. J Ovarian Res 2013; 6: 95.
Steeg PS . Targeting metastasis. Nat Rev Cancer 2016; 16: 201–218.
Beekman KW, Colevas AD, Cooney K, Dipaola R, Dunn RL, Gross M et al. Phase II evaluations of cilengitide in asymptomatic patients with androgen-independent prostate cancer: scientific rationale and study design. Clin Genitourin Cancer 2006; 4: 299–302.
Reardon DA, Fink KL, Mikkelsen T, Cloughesy TF, O'Neill A, Plotkin S et al. Randomized phase II study of cilengitide, an integrin-targeting arginine-glycine-aspartic acid peptide, in recurrent glioblastoma multiforme. J Clin Oncol 2008; 26: 5610–5617.
Reynolds AR, Hart IR, Watson AR, Welti JC, Silva RG, Robinson SD et al. Stimulation of tumor growth and angiogenesis by low concentrations of RGD-mimetic integrin inhibitors. Nat Med 2009; 15: 392–400.
Li G, Stahl PD . Post-translational processing and membrane association of the two early endosome-associated rab GTP-binding proteins (rab4 and rab5). Arch Biochem Biophys 1993; 304: 471–478.
Knight JB, Cao KT, Gibson GV, Olson AL . Expression of a prenylation-deficient Rab4 interferes with propagation of insulin signaling through insulin receptor substrate-1. Endocrinology 2000; 141: 208–218.
Manu KA, Chai TF, Teh JT, Zhu WL, Casey PJ, Wang M . Inhibition of isoprenylcysteine carboxylmethyltransferase induces cell-cycle arrest and apoptosis through p21 and p21-regulated BNIP3 induction in pancreatic cancer. Mol Cancer Ther 2017; 16: 914–923.
Ballestrem C, Hinz B, Imhof BA, Wehrle-Haller B . Marching at the front and dragging behind: differential alphaVbeta3-integrin turnover regulates focal adhesion behavior. J Cell Biol 2001; 155: 1319–1332.
Kutner RH, Zhang XY, Reiser J . Production, concentration and titration of pseudotyped HIV-1-based lentiviral vectors. Nat Protoc 2009; 4: 495–505.
Zhu WL, Hossain MS, Guo DY, Liu S, Tong H, Khakpoor A et al. A role for Rac3 GTPase in the regulation of autophagy. J Biol Chem 2011; 286: 35291–35298.
Acknowledgements
This work is supported by Singapore Ministries of Education and Ministry of Health for funding of this project through their Tier II and CSA schemes, respectively. We are grateful for the help on CAM assays by Dr Suhail Ahmed Kabeer Rasheed of Duke-NUS. The authors confirm that neither the submitted manuscript nor any similar manuscript, in whole or in part, is under consideration, in press, or reported elsewhere.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Oncogene website
Supplementary information
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/4.0/
About this article
Cite this article
Do, M., Chai, T., Casey, P. et al. Isoprenylcysteine carboxylmethyltransferase function is essential for RAB4A-mediated integrin β3 recycling, cell migration and cancer metastasis. Oncogene 36, 5757–5767 (2017). https://doi.org/10.1038/onc.2017.183
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/onc.2017.183
This article is cited by
-
Protein lipidation in cancer: mechanisms, dysregulation and emerging drug targets
Nature Reviews Cancer (2024)
-
RAB4A GTPase regulates epithelial-to-mesenchymal transition by modulating RAC1 activation
Breast Cancer Research (2022)
-
Endocytosis: a pivotal pathway for regulating metastasis
British Journal of Cancer (2021)
-
A SNAI2-PEAK1-INHBA stromal axis drives progression and lapatinib resistance in HER2-positive breast cancer by supporting subpopulations of tumor cells positive for antiapoptotic and stress signaling markers
Oncogene (2021)
-
Isoprenylcysteine carboxylmethyltransferase is required for the impact of mutant KRAS on TAZ protein level and cancer cell self-renewal
Oncogene (2020)