Abstract
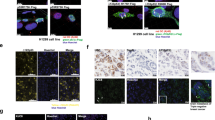
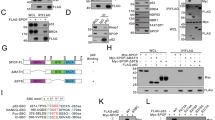
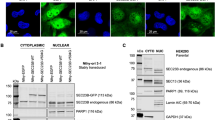
Mutation of the TP53 gene represents a prevalent genetic alteration in human cancers, and a subset of p53 mutants may form amyloid-like aggregates that contribute to the gain of oncogenic functions (GOFs) and chemoresistance. Here we identify the pathways that may mediate the aggregation-associated GOF by using combined proteomic analysis and genome-wide recruitment profiling. Mass spectrometry revealed activation of unfolded protein response (UPR) pathway and upregulation of endoplasmic reticulum protein 29 (ERp29) in R282WTP53-expressing cells that were exposed to cisplatin stress. Chromatin immunoprecipitation sequencing identified a significant 'CCCASS' binding motif of Arg282Trp, which is present in the promoter region of ERP29 gene. The mutant p53 upregulated ERP29 mRNA and protein expression levels, whereas targeting ERP29 by specific small interfering RNAs suppressed the chemoresistant effect of Arg282Trp. The anti-aggregation peptide ReACp53 significantly decreased ERP29 expression and suppressed the chemoresistant effect. These findings highlight a role of ERP29 in the acquired chemoresistance of cancer cells expressing the aggregating p53 mutant Arg282Trp. Our results also suggest that ERP29-mediated GOF can be targeted by the anti-aggregation peptide ReACp53.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Acin S, Li Z, Mejia O, Roop DR, El-Naggar AK, Caulin C . Gain-of-function mutant p53 but not p53 deletion promotes head and neck cancer progression in response to oncogenic K-ras. J Pathol 2011; 225: 479–489.
Al-Rawashdeh FY, Scriven P, Cameron IC, Vergani PV, Wyld L . Unfolded protein response activation contributes to chemoresistance in hepatocellular carcinoma. Eur J Gastroenterol Hepatol 2010; 22: 1099–1105.
Ali A, Shah AS, Ahmad A . Gain-of-function of mutant p53: mutant p53 enhances cancer progression by inhibiting KLF17 expression in invasive breast carcinoma cells. Cancer Lett 2014; 354: 87–96.
Ano Bom AP, Rangel LP, Costa DC, de Oliveira GA, Sanches D, Braga CA et al. Mutant p53 aggregates into prion-like amyloid oligomers and fibrils: implications for cancer. J Biol Chem 2012; 287: 28152–28162.
Bailey TL . DREME: motif discovery in transcription factor ChIP-seq data. Bioinformatics 2011; 27: 1653–1659.
Brosh R, Rotter V . When mutants gain new powers: news from the mutant p53 field. Nat Rev Cancer 2009; 9: 701–713.
Chen S, Zhang D . Friend or foe: endoplasmic reticulum protein 29 (ERp29) in epithelial cancer. FEBS Open Bio 2015; 5: 91–98.
Chen S, Zhang Y, Zhang D . Endoplasmic reticulum protein 29 (ERp29) confers radioresistance through the DNA repair gene, O(6)-methylguanine DNA-methyltransferase, in breast cancer cells. Sci Rep 2015; 5: 14723.
Cheung EC, Vousden KH . The role of p53 in glucose metabolism. Curr Opin Cell Biol 2010; 22: 186–191.
Chiang MF, Yeh ST, Liao HF, Chang NS, Chen YJ . Overexpression of WW domain-containing oxidoreductase WOX1 preferentially induces apoptosis in human glioblastoma cells harboring mutant p53. Biomed Pharmacother 2012; 66: 433–438.
Cino EA, Soares IN, Pedrote MM, de Oliveira GA, Silva JL . Aggregation tendencies in the p53 family are modulated by backbone hydrogen bonds. Sci Rep 2016; 6: 32535.
Coffill CR, Muller PA, Oh HK, Neo SP, Hogue KA, Cheok CF et al. Mutant p53 interactome identifies nardilysin as a p53R273H-specific binding partner that promotes invasion. EMBO Rep 2012; 13: 638–644.
Das S, Smith TD, Sarma JD, Ritzenthaler JD, Maza J, Kaplan BE et al. ERp29 restricts Connexin43 oligomerization in the endoplasmic reticulum. Mol Biol Cell 2009; 20: 2593–2604.
Datta A, Dey S, Das P, Alam SK, Roychoudhury S . Transcriptome profiling identifies genes and pathways deregulated upon floxuridine treatment in colorectal cancer cells harboring GOF mutant p53. Genomics Data 2016; 8: 47–51.
Di Fiore R, Marcatti M, Drago-Ferrante R, D'Anneo A, Giuliano M, Carlisi D et al. Mutant p53 gain of function can be at the root of dedifferentiation of human osteosarcoma MG63 cells into 3AB-OS cancer stem cells. Bone 2014; 60: 198–212.
Do PM, Varanasi L, Fan S, Li C, Kubacka I, Newman V et al. Mutant p53 cooperates with ETS2 to promote etoposide resistance. Genes Dev 2012; 26: 830–845.
Dudgeon C, Chan C, Kang W, Sun Y, Emerson R, Robins H et al. The evolution of thymic lymphomas in p53 knockout mice. Genes Dev 2014; 28: 2613–2620.
Epple LM, Dodd RD, Merz AL, Dechkovskaia AM, Herring M, Winston BA et al. Induction of the unfolded protein response drives enhanced metabolism and chemoresistance in glioma cells. PLoS ONE 2013; 8: e73267.
Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer 2015; 136: E359–E386.
Freed-Pastor WA, Prives C . Mutant p53: one name, many proteins. Genes Dev 2012; 26: 1268–1286.
Gifford JB, Huang W, Zeleniak AE, Hindoyan A, Wu H, Donahue TR et al. Expression of GRP78, master regulator of the unfolded protein response, increases chemoresistance in pancreatic ductal adenocarcinoma. Mol Cancer Ther 2016; 15: 1043–1052.
Guan Z, Wang XR, Zhu XF, Huang XF, Xu J, Wang LH et al. Aurora-A, a negative prognostic marker, increases migration and decreases radiosensitivity in cancer cells. Cancer Res 2007; 67: 10436–10444.
He C, Li L, Guan X, Xiong L, Miao X . Mutant p53 gain of function and chemoresistance: the role of mutant p53 in response to clinical chemotherapy. Chemotherapy 2016; 62: 43–53.
Hirsch I, Weiwad M, Prell E, Ferrari DM . ERp29 deficiency affects sensitivity to apoptosis via impairment of the ATF6-CHOP pathway of stress response. Apoptosis 2014; 19: 801–815.
Hume MA, Barrera LA, Gisselbrecht SS, Bulyk ML . UniPROBE update 2015: new tools and content for the online database of protein-binding microarray data on protein-DNA interactions. Nucleic Acids Res 2015; 43: D117–D122.
Humpton TJ, Vousden KH . Regulation of cellular metabolism and hypoxia by p53. Cold Spring Harb Perspect Med 2016; 6: a026146.
Kim E, Deppert W . Interactions of mutant p53 with DNA: guilt by association. Oncogene 2007; 26: 2185–2190.
Kim MK, Cho JH, Lee JJ, Son MH, Lee KJ . Proteomic analysis of INS-1 rat insulinoma cells: ER stress effects and the protective role of exenatide, a GLP-1 receptor agonist. PLoS ONE 2015; 10: e0120536.
Kiprijanovska S, Stavridis S, Stankov O, Komina S, Petrusevska G, Polenakovic M et al. Mapping and identification of the urine proteome of prostate cancer patients by 2D PAGE/MS. Int J Proteomics 2014; 2014: 594761.
Krishnan SR, Nair BC, Sareddy GR, Roy SS, Natarajan M, Suzuki T et al. Novel role of PELP1 in regulating chemotherapy response in mutant p53-expressing triple negative breast cancer cells. Breast Cancer Res Treat 2015; 150: 487–499.
Lasagna-Reeves CA, Clos AL, Castillo-Carranza D, Sengupta U, Guerrero-Munoz M, Kelly B et al. Dual role of p53 amyloid formation in cancer; loss of function and gain of toxicity. Biochem Biophys Res Commun 2013; 430: 963–968.
Lavra L, Ulivieri A, Rinaldo C, Dominici R, Volante M, Luciani E et al. Gal-3 is stimulated by gain-of-function p53 mutations and modulates chemoresistance in anaplastic thyroid carcinomas. J Pathol 2009; 218: 66–75.
Lee E, Nichols P, Spicer D, Groshen S, Yu MC, Lee AS . GRP78 as a novel predictor of responsiveness to chemotherapy in breast cancer. Cancer Res 2006; 66: 7849–7853.
Levy CB, Stumbo AC, Ano Bom AP, Portari EA, Cordeiro Y, Silva JL et al. Co-localization of mutant p53 and amyloid-like protein aggregates in breast tumors. Int J Biochem Cell Biol 2011; 43: 60–64.
Li D, Yallowitz A, Ozog L, Marchenko N . A gain-of-function mutant p53-HSF1 feed forward circuit governs adaptation of cancer cells to proteotoxic stress. Cell Death Dis 2014; 5: e1194.
Lu JJ, Lu DZ, Chen YF, Dong YT, Zhang JR, Li T et al. Proteomic analysis of hepatocellular carcinoma HepG2 cells treated with platycodin D. Chin J Nat Med 2015; 13: 673–679.
Maere S, Heymans K, Kuiper M . BiNGO: a cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics 2005; 21: 3448–3449.
Magdeldin S, Enany S, Yoshida Y, Xu B, Zhang Y, Zureena Z et al. Basics and recent advances of two dimensional- polyacrylamide gel electrophoresis. Clin Proteomics 2014; 11: 16.
Mantovani F, Walerych D, Sal GD . Targeting mutant p53 in cancer: a long road to precision therapy. FEBS J 2016; 284: 837–850.
Mathelier A, Fornes O, Arenillas DJ, Chen CY, Denay G, Lee J et al. JASPAR 2016: a major expansion and update of the open-access database of transcription factor binding profiles. Nucleic Acids Res 2016; 44: D110–D115.
May C, Brosseron F, Pfeiffer K, Meyer HE, Marcus K . Proteome analysis with classical 2D-PAGE. Methods Mol Biol 2012; 893: 37–46.
Mori-Iwamoto S, Kuramitsu Y, Ryozawa S, Mikuria K, Fujimoto M, Maehara S et al. Proteomics finding heat shock protein 27 as a biomarker for resistance of pancreatic cancer cells to gemcitabine. Int J Oncol 2007; 31: 1345–1350.
Muller PA, Vousden KH . p53 mutations in cancer. Nat Cell Biol 2013; 15: 2–8.
Neilsen PM, Noll JE, Suetani RJ, Schulz RB, Al-Ejeh F, Evdokiou A et al. Mutant p53 uses p63 as a molecular chaperone to alter gene expression and induce a pro-invasive secretome. Oncotarget 2011; 2: 1203–1217.
Nesslinger NJ, Shi XB, deVere White RW . Androgen-independent growth of LNCaP prostate cancer cells is mediated by gain-of-function mutant p53. Cancer Res 2003; 63: 2228–2233.
Oren M, Rotter V . Mutant p53 gain-of-function in cancer. Cold Spring Harb Perspect Biol 2010; 2: a001107.
Pi L, Li X, Song Q, Shen Y, Lu X, Di B . Knockdown of glucose-regulated protein 78 abrogates chemoresistance of hypopharyngeal carcinoma cells to cisplatin induced by unfolded protein in response to severe hypoxia. Oncol Lett 2014; 7: 685–692.
Polotskaia A, Xiao G, Reynoso K, Martin C, Qiu WG, Hendrickson RC et al. Proteome-wide analysis of mutant p53 targets in breast cancer identifies new levels of gain-of-function that influence PARP, PCNA, and MCM4. Proc Natl Acad Sci USA 2015; 112: E1220–E1229.
Qi L, Wu P, Zhang X, Qiu Y, Jiang W, Huang D et al. Inhibiting ERp29 expression enhances radiosensitivity in human nasopharyngeal carcinoma cell lines. Med Oncol 2012; 29: 721–728.
Schwartz M, Zhang Y, Rosenblatt JD . B cell regulation of the anti-tumor response and role in carcinogenesis. J Immunother Cancer 2016; 4: 40.
Silva JL, De Moura Gallo CV, Costa DC, Rangel LP . Prion-like aggregation of mutant p53 in cancer. Trends Biochem Sci 2014; 39: 260–267.
Song IK, Lee JJ, Cho JH, Jeong J, Shin DH, Lee KJ . Degradation of redox-sensitive proteins including peroxiredoxins and DJ-1 is promoted by oxidation-induced conformational changes and ubiquitination. Sci Rep 2016; 6: 34432.
Soragni A, Janzen DM, Johnson LM, Lindgren AG, Thai-Quynh Nguyen A, Tiourin E et al. A designed inhibitor of p53 aggregation rescues p53 tumor suppression in ovarian carcinomas. Cancer Cell 2016; 29: 90–103.
Vaughan CA, Frum R, Pearsall I, Singh S, Windle B, Yeudall A et al. Allele specific gain-of-function activity of p53 mutants in lung cancer cells. Biochem Biophys Res Commun 2012; 428: 6–10.
Vousden KH, Ryan KM . p53 and metabolism. Nat Rev Cancer 2009; 9: 691–700.
Walerych D, Lisek K, Del Sal G . Mutant p53: one, no one, and one hundred thousand. Front Oncol 2015; 5: 289.
Walerych D, Lisek K, Del Sal G . Multi-omics reveals global effects of mutant p53 gain-of-function. Cell cycle 2016; 15: 3009–3010.
Walerych D, Lisek K, Sommaggio R, Piazza S, Ciani Y, Dalla E et al. Proteasome machinery is instrumental in a common gain-of-function program of the p53 missense mutants in cancer. Nat Cell Biol 2016; 18: 897–909.
Wang X, Chen JX, Liu JP, You C, Liu YH, Mao Q . Gain of function of mutant TP53 in glioblastoma: prognosis and response to temozolomide. Ann Surg Oncol 2014; 21: 1337–1344.
Xu J, Reumers J, Couceiro JR, De Smet F, Gallardo R, Rudyak S et al. Gain of function of mutant p53 by coaggregation with multiple tumor suppressors. Nat Chem Biol 2011; 7: 285–295.
Xu J, Qian J, Hu Y, Wang J, Zhou X, Chen H et al. Heterogeneity of Li-Fraumeni syndrome links to unequal gain-of-function effects of p53 mutations. Sci Rep 2014; 4: 4223.
Xu J, Wang J, Hu Y, Qian J, Xu B, Chen H et al. Unequal prognostic potentials of p53 gain-of-function mutations in human cancers associate with drug-metabolizing activity. Cell Death Dis 2014; 5: e1108.
Yang-Hartwich Y, Soteras MG, Lin ZP, Holmberg J, Sumi N, Craveiro V et al. p53 protein aggregation promotes platinum resistance in ovarian cancer. Oncogene 2015; 34: 3605–3616.
Zhang D, Putti TCx . Over-expression of ERp29 attenuates doxorubicin-induced cell apoptosis through up-regulation of Hsp27 in breast cancer cells. Exp Cell Res 2010; 316: 3522–3531.
Zhang Y, Hu Y, Fang JY, Xu J . Gain-of-function miRNA signature by mutant p53 associates with poor cancer outcome. Oncotarget 2016; 7: 11056–11066.
Zhou G, Wang J, Zhao M, Xie TX, Tanaka N, Sano D et al. Gain-of-function mutant p53 promotes cell growth and cancer cell metabolism via inhibition of AMPK activation. Mol Cell 2014; 54: 960–974.
Acknowledgements
This project was supported by grants from National Natural Science Foundation of China (81572326, 81322036, 81272383, 81421001, 81320108024, 81530072, 81602518, 81502015 and 81572303); Top-Notch Young Talents Program of China (ZTZ2015–48); Shanghai Municipal Education Commission–Gaofeng Clinical Medicine Grant Support (20152514); National Key Research & Development (R&D) Plan (2016YFC0906000 and 2016YFC0906002); and National Key Technology Support Program (2015BAI13B07). The sponsors of this study had no role in the collection of the data, the analysis and interpretation of the data, the decision to submit the manuscript for publication or the writing of the manuscript.
Author contributions
YZ and JX wrote the manuscript. YZ, YH, J-LW, HY, HW, LL, CL, HS, YC and J-YF performed experiments and/or analyzed the results.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Oncogene website
Supplementary information
Rights and permissions
About this article
Cite this article
Zhang, Y., Hu, Y., Wang, JL. et al. Proteomic identification of ERP29 as a key chemoresistant factor activated by the aggregating p53 mutant Arg282Trp. Oncogene 36, 5473–5483 (2017). https://doi.org/10.1038/onc.2017.152
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/onc.2017.152
This article is cited by
-
Targeting p53 pathways: mechanisms, structures, and advances in therapy
Signal Transduction and Targeted Therapy (2023)
-
Potential of rescue and reactivation of tumor suppressor p53 for cancer therapy
Biophysical Reviews (2022)
-
Therapeutic potential of ReACp53 targeting mutant p53 protein in CRPC
Prostate Cancer and Prostatic Diseases (2020)
-
Role of a genetic variation in the microRNA-4421 binding site of ERP29 regarding risk of oropharynx cancer and prognosis
Scientific Reports (2020)
-
HIP1R targets PD-L1 to lysosomal degradation to alter T cell–mediated cytotoxicity
Nature Chemical Biology (2019)