Abstract
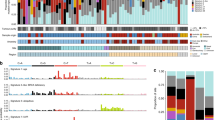
Despite remarkable progress in cutaneous melanoma genomic profiling, the mutational landscape of primary mucosal melanomas (PMM) remains unclear. Forty-six PMMs underwent targeted exome sequencing of 111 cancer-associated genes. Seventy-six somatic nonsynonymous mutations in 42 genes were observed, and recurrent mutations were noted on eight genes, including TP53 (13%), NRAS (13%), SNX31 (9%), NF1 (9%), KIT (7%) and APC (7%). Mitogen-activated protein kinase (MAPK; 37%), cell cycle (20%) and phosphatidylinositol 3-kinase (PI3K)-mTOR (15%) pathways were frequently mutated. We biologically characterized a novel ZNF767-BRAF fusion found in a vemurafenib-refractory respiratory tract PMM, from which cell line harboring ZNF767-BRAF fusion were established for further molecular analyses. In an independent data set, NFIC-BRAF fusion was identified in an oral PMM case and TMEM178B-BRAF fusion and DGKI-BRAF fusion were identified in two malignant melanomas with a low mutational burden (number of mutation per megabase, 0.8 and 4, respectively). Subsequent analyses revealed that the ZNF767-BRAF fusion protein promotes RAF dimerization and activation of the MAPK pathway. We next tested the in vitro and in vivo efficacy of vemurafenib, trametinib, BKM120 or LEE011 alone and in combination. Trametinib effectively inhibited tumor cell growth in vitro, but the combination of trametinib and BKM120 or LEE011 yielded more than additive anti-tumor effects both in vitro and in vivo in a melanoma cells harboring the BRAF fusion. In conclusion, BRAF fusions define a new molecular subset of PMM that can be targeted therapeutically by the combination of a MEK inhibitor with PI3K or cyclin-dependent kinase 4/6 inhibitors.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Chang AE, Karnell LH, Menck HR . The National Cancer Data Base report on cutaneous and noncutaneous melanoma: a summary of 84836 cases from the past decade. The American College of Surgeons Commission on Cancer and the American Cancer Society. Cancer 1998; 83: 1664–1678.
Mihajlovic M, Vlajkovic S, Jovanovic P, Stefanovic V . Primary mucosal melanomas: a comprehensive review. Int J Clin Exp Pathol 2012; 5: 739–753.
Seetharamu N, Ott PA, Pavlick AC . Mucosal melanomas: a case-based review of the literature. Oncologist 2010; 15: 772–781.
National Comprehensive Cancer Network (NCCN). NCCN Clinical Practice Guidelines in Oncology: Melanoma. Version 1. 2017. Available at:https://www.nccn.org/.
Gray-Schopfer V, Wellbrock C, Marais R . Melanoma biology and new targeted therapy. Nature 2007; 445: 851–857.
Sullivan RJ, Flaherty K . MAP kinase signaling and inhibition in melanoma. Oncogene 2013; 32: 2373–2379.
Wellbrock C, Ogilvie L, Hedley D, Karasarides M, Martin J, Niculescu-Duvaz D et al. V599EB-RAF is an oncogene in melanocytes. Cancer Res 2004; 64: 2338–2342.
Campbell PM, Der CJ . Oncogenic Ras and its role in tumor cell invasion and metastasis. Semin Cancer Biol 2004; 14: 105–114.
Curtin JA, Busam K, Pinkel D, Bastian BC . Somatic activation of KIT in distinct subtypes of melanoma. J Clin Oncol 2006; 24: 4340–4346.
Flaherty KT, Puzanov I, Kim KB, Ribas A, McArthur GA, Sosman JA et al. Inhibition of mutated, activated BRAF in metastatic melanoma. N Engl J Med 2010; 363: 809–819.
Hodi FS, O’Day SJ, McDermott DF, Weber RW, Sosman JA, Haanen JB et al. Improved survival with ipilimumab in patients with metastatic melanoma. N Engl J Med 2010; 363: 711–723.
Hamid O, Robert C, Daud A, Hodi FS, Hwu WJ, Kefford R et al. Safety and tumor responses with lambrolizumab (anti-PD-1) in melanoma. N Engl J Med 2013; 369: 134–144.
Robert C, Karaszewska B, Schachter J, Rutkowski P, Mackiewicz A, Stroiakovski D et al. Improved overall survival in melanoma with combined dabrafenib and trametinib. N Engl J Med 2015; 372: 30–39.
Deuker MM, Marsh Durban V, Phillips WA, McMahon M . PI3'-kinase inhibition forestalls the onset of MEK1/2 inhibitor resistance in BRAF-mutated melanoma. Cancer Discov 2015; 5: 143–153.
Kwong LN, Costello JC, Liu H, Jiang S, Helms TL, Langsdorf AE et al. Oncogenic NRAS signaling differentially regulates survival and proliferation in melanoma. Nat Med 2012; 18: 1503–1510.
Ross JS, Wang K, Chmielecki J, Gay L, Johnson A, Chudnovsky J et al. The distribution of BRAF gene fusions in solid tumors and response to targeted therapy. Int J Cancer 2016; 138: 881–890.
McVey M, Lee SE . MMEJ repair of double-strand breaks (director’s cut): deleted sequences and alternative endings. Trends Genet 2008; 24: 529–538.
Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz Jr LA, Kinzler KW . Cancer genome landscapes. Science 2013; 339: 1546–1558.
Cancer Genome Atlas Network. Electronic address IMO, Cancer Genome Atlas N. Genomic classification of cutaneous melanoma. Cell 2015; 161: 1681–1696.
Cutler Jr RE, Stephens RM, Saracino MR, Morrison DK . Autoregulation of the Raf-1 serine/threonine kinase. Proc Natl Acad Sci USA 1998; 95: 9214–9219.
Flaherty KT, Robert C, Hersey P, Nathan P, Garbe C, Milhem M et al. Improved survival with MEK inhibition in BRAF-mutated melanoma. N Engl J Med 2012; 367: 107–114.
Chapman PB, Hauschild A, Robert C, Haanen JB, Ascierto P, Larkin J et al. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N Engl J Med 2011; 364: 2507–2516.
Zhang C, Spevak W, Zhang Y, Burton EA, Ma Y, Habets G et al. RAF inhibitors that evade paradoxical MAPK pathway activation. Nature 2015; 526: 583–586.
Girotti MR, Lopes F, Preece N, Niculescu-Duvaz D, Zambon A, Davies L et al. Paradox-breaking RAF inhibitors that also target SRC are effective in drug-resistant BRAF mutant melanoma. Cancer Cell 2015; 27: 85–96.
Posch C, Moslehi H, Feeney L, Green GA, Ebaee A, Feichtenschlager V et al. Combined targeting of MEK and PI3K/mTOR effector pathways is necessary to effectively inhibit NRAS mutant melanomain vitro and in vivo. Proc Natl Acad Sci USA 2013; 110: 4015–4020.
Hutchinson KE, Lipson D, Stephens PJ, Otto G, Lehmann BD, Lyle PL et al. BRAF fusions define a distinct molecular subset of melanomas with potential sensitivity to MEK inhibition. Clin Cancer Res 2013; 19: 6696–6702.
Palanisamy N, Ateeq B, Kalyana-Sundaram S, Pflueger D, Ramnarayanan K, Shankar S et al. Rearrangements of the RAF kinase pathway in prostate cancer, gastric cancer and melanoma. Nat Med 2010; 16: 793–798.
Ciampi R, Knauf JA, Kerler R, Gandhi M, Zhu Z, Nikiforova MN et al. Oncogenic AKAP9-BRAF fusion is a novel mechanism of MAPK pathway activation in thyroid cancer. J Clin Invest 2005; 115: 94–101.
Jeuken JW, Wesseling P . MAPK pathway activation through BRAF gene fusion in pilocytic astrocytomas; a novel oncogenic fusion gene with diagnostic, prognostic, and therapeutic potential. J Pathol 2010; 222: 324–328.
Dessars B, De Raeve LE, El Housni H, Debouck CJ, Sidon PJ, Morandini R et al. Chromosomal translocations as a mechanism of BRAF activation in two cases of large congenital melanocytic nevi. J Invest Dermatol 2007; 127: 1468–1470.
Forbes SA, Beare D, Gunasekaran P, Leung K, Bindal N, Boutselakis H et al. COSMIC: exploring the world’s knowledge of somatic mutations in human cancer. Nucleic Acids Res 2015; 43: D805–D811.
Rajakulendran T, Sahmi M, Lefrancois M, Sicheri F, Therrien M . A dimerization-dependent mechanism drives RAF catalytic activation. Nature 2009; 461: 542–545.
Poulikakos PI, Persaud Y, Janakiraman M, Kong X, Ng C, Moriceau G et al. RAF inhibitor resistance is mediated by dimerization of aberrantly spliced BRAF(V600E). Nature 2011; 480: 387–390.
Worm J, Christensen C, Gronbaek K, Tulchinsky E, Guldberg P . Genetic and epigenetic alterations of the APC gene in malignant melanoma. Oncogene 2004; 23: 5215–5226.
Hodis E, Watson IR, Kryukov GV, Arold ST, Imielinski M, Theurillat JP et al. A landscape of driver mutations in melanoma. Cell 2012; 150: 251–263.
Ghai R, Mobli M, Norwood SJ, Bugarcic A, Teasdale RD, King GF et al. Phox homology band 4.1/ezrin/radixin/moesin-like proteins function as molecular scaffolds that interact with cargo receptors and Ras GTPases. Proc Natl Acad Sci USA 2011; 108: 7763–7768.
Giehl K . Oncogenic Ras in tumour progression and metastasis. Biol Chem 2005; 386: 193–205.
Stark MS, Woods SL, Gartside MG, Bonazzi VF, Dutton-Regester K, Aoude LG et al. Frequent somatic mutations in MAP3K5 and MAP3K9 in metastatic melanoma identified by exome sequencing. Nat Genet 2012; 44: 165–169.
Nikolaev SI, Rimoldi D, Iseli C, Valsesia A, Robyr D, Gehrig C et al. Exome sequencing identifies recurrent somatic MAP2K1 and MAP2K2 mutations in melanoma. Nat Genet 2012; 44: 133–139.
Krauthammer M, Kong Y, Ha BH, Evans P, Bacchiocchi A, McCusker JP et al. Exome sequencing identifies recurrent somatic RAC1 mutations in melanoma. Nat Genet 2012; 44: 1006–1014.
Van Allen EM, Wagle N, Stojanov P, Perrin DL, Cibulskis K, Marlow S et al. Whole-exome sequencing and clinical interpretation of formalin-fixed, paraffin-embedded tumor samples to guide precision cancer medicine. Nat Med 2014; 20: 682–688.
Li H, Durbin R . Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 2010; 26: 589–595.
DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet 2011; 43: 491–498.
Cibulskis K, Lawrence MS, Carter SL, Sivachenko A, Jaffe D, Sougnez C et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol 2013; 31: 213–219.
Koboldt DC, Zhang Q, Larson DE, Shen D, McLellan MD, Lin L et al. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res 2012; 22: 568–576.
Wang K, Li M, Hakonarson H . ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 2010; 38: e164.
Yang Y, Muzny DM, Reid JG, Bainbridge MN, Willis A, Ward PA et al. Clinical whole-exome sequencing for the diagnosis of mendelian disorders. N Engl J Med 2013; 369: 1502–1511.
Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL . TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 2013; 14: R36.
Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ et al. Transcript assembly and quantification by RNA-seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 2010; 28: 511–515.
Iyer MK, Chinnaiyan AM, Maher CA . ChimeraScan: a tool for identifying chimeric transcription in sequencing data. Bioinformatics 2011; 27: 2903–2904.
Ge H, Liu K, Juan T, Fang F, Newman M, Hoeck W . FusionMap: detecting fusion genes from next-generation sequencing data at base-pair resolution. Bioinformatics 2011; 27: 1922–1928.
Chou TC . Drug combination studies and their synergy quantification using the Chou-Talalay method. Cancer Res 2010; 70: 440–446.
Acknowledgements
This study was supported in part by a grant from the Basic Science Research Program through the National Research Foundation of Korea (NRF), funded by the Ministry of Science, ICT and Future Planning (2014R1A1A1004823 to M Jung), and the NRF by the Korean government (MEST; 2012R1A2A2A01046927 to B Cho). The results of the analysis of mutation frequencies in cutaneous melanomas compared with corresponding frequencies in PMM were based on TCGA Research Network generated data (http://cancergenome.nih.gov/).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Oncogene website http://www.nature.com/onc.
Supplementary information
Rights and permissions
About this article
Cite this article
Kim, H., Jung, M., Kang, H. et al. Oncogenic BRAF fusions in mucosal melanomas activate the MAPK pathway and are sensitive to MEK/PI3K inhibition or MEK/CDK4/6 inhibition. Oncogene 36, 3334–3345 (2017). https://doi.org/10.1038/onc.2016.486
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/onc.2016.486
This article is cited by
-
Evolving Treatment Approaches to Mucosal Melanoma
Current Oncology Reports (2022)
-
Alterations in key signaling pathways in sinonasal tract melanoma. A molecular genetics and immunohistochemical study of 90 cases and comprehensive review of the literature
Modern Pathology (2022)
-
Genetic alteration of Chinese patients with rectal mucosal melanoma
BMC Cancer (2021)
-
Kinase gene fusions: roles and therapeutic value in progressive and refractory papillary thyroid cancer
Journal of Cancer Research and Clinical Oncology (2021)
-
ALK detection in lung cancer: identification of atypical and cryptic ALK rearrangements using an optimal algorithm
Journal of Cancer Research and Clinical Oncology (2020)