Abstract
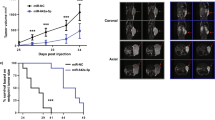
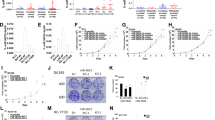
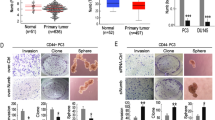
A major genomic alteration in prostate cancer (PCa) is frequent loss of chromosome (chr) 8p with a common region of loss of heterozygosity (LOH) at chr8p22 locus. Genomic studies implicate this locus in the initiation of clinically significant PCa and with progression to metastatic disease. However, the genes within this region have not been fully characterized to date. Here we demonstrate for the first time that a microRNA component of this region—miR-383—is frequently downregulated in prostate cancer, has a critical role in determining tumor-initiating potential and is involved in prostate cancer metastasis via direct regulation of CD44, a ubiquitous marker of PCa tumor-initiating cells (TICs)/stem cells. Expression analyses of miR-383 in PCa clinical tissues established that low miR-383 expression is associated with poor prognosis. Functional data suggest that miR-383 regulates PCa tumor-initiating/stem-like cells via CD44 regulation. Ectopic expression of miR-383 inhibited tumor-initiating capacity of CD44+ PCa cells. Also, ‘anti-metastatic’ effects of ectopic miR-383 expression were observed in a PCa experimental metastasis model. In view of our results, we propose that frequent loss of miR-383 at chr8p22 region leads to tumor initiation and prostate cancer metastasis. Thus, we have identified a novel finding that associates a long observed genomic alteration to PCa stemness and metastasis. Our data suggest that restoration of miR-383 expression may be an effective therapeutic modality against PCa. Importantly, we identified miR-383 as a novel PCa tissue diagnostic biomarker with a potential that outperforms that of serum PSA.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
References
Tang DG, Patrawala L, Calhoun T, Bhatia B, Choy G, Schneider-Broussard R et al. Prostate cancer stem/progenitor cells: identification, characterization, and implications. Mol Carcinog 2007; 46: 1–14.
Shen MM, Abate-Shen C . Molecular genetics of prostate cancer: new prospects for old challenges. Genes Dev 2010; 24: 1967–2000.
Li H, Tang DG . Prostate cancer stem cells and their potential roles in metastasis. J Surg Oncol 2011; 103: 558–562.
Wicha MS . Stemming a tumor with a little miR. Nat Med 2011; 17: 162–164.
Collins AT, Berry PA, Hyde C, Stower MJ, Maitland NJ . Prospective identification of tumorigenic prostate cancer stem cells. Cancer Res 2005; 65: 10946–10951.
Li H, Chen X, Calhoun-Davis T, Claypool K, Tang DG . PC3 human prostate carcinoma cell holoclones contain self-renewing tumor-initiating cells. Cancer Res 2008; 68: 1820–1825.
Patrawala L, Calhoun T, Schneider-Broussard R, Li H, Bhatia B, Tang S et al. Highly purified CD44+ prostate cancer cells from xenograft human tumors are enriched in tumorigenic and metastatic progenitor cells. Oncogene 2006; 25: 1696–1708.
Patrawala L, Calhoun T, Schneider-Broussard R, Zhou J, Claypool K, Tang DG . Side population is enriched in tumorigenic, stem-like cancer cells, whereas ABCG2+ and ABCG2- cancer cells are similarly tumorigenic. Cancer Res 2005; 65: 6207–6219.
Patrawala L, Calhoun-Davis T, Schneider-Broussard R, Tang DG . Hierarchical organization of prostate cancer cells in xenograft tumors: the CD44+alpha2beta1+ cell population is enriched in tumor-initiating cells. Cancer Res 2007; 67: 6796–6805.
Iczkowski KA . Cell adhesion molecule CD44: its functional roles in prostate cancer. Am J Transl Res 2010; 3: 1–7.
Kim JH, Dhanasekaran SM, Mehra R, Tomlins SA, Gu W, Yu J et al. Integrative analysis of genomic aberrations associated with prostate cancer progression. Cancer Res 2007; 67: 8229–8239.
Lapointe J, Li C, Giacomini CP, Salari K, Huang S, Wang P et al. Genomic profiling reveals alternative genetic pathways of prostate tumorigenesis. Cancer Res 2007; 67: 8504–8510.
Lapointe J, Li C, Higgins JP, van de Rijn M, Bair E, Montgomery K et al. Gene expression profiling identifies clinically relevant subtypes of prostate cancer. Proc Natl Acad Sci USA 2004; 101: 811–816.
Lieberfarb ME, Lin M, Lechpammer M, Li C, Tanenbaum DM, Febbo PG et al. Genome-wide loss of heterozygosity analysis from laser capture microdissected prostate cancer using single nucleotide polymorphic allele (SNP) arrays and a novel bioinformatics platform dChipSNP. Cancer Res 2003; 63: 4781–4785.
Perner S, Demichelis F, Beroukhim R, Schmidt FH, Mosquera JM, Setlur S et al. TMPRSS2:ERG fusion-associated deletions provide insight into the heterogeneity of prostate cancer. Cancer Res 2006; 66: 8337–8341.
Singh D, Febbo PG, Ross K, Jackson DG, Manola J, Ladd C et al. Gene expression correlates of clinical prostate cancer behavior. Cancer Cell 2002; 1: 203–209.
Taylor BS, Schultz N, Hieronymus H, Gopalan A, Xiao Y, Carver BS et al. Integrative genomic profiling of human prostate cancer. Cancer Cell 2010; 18: 11–22.
Bova GS, Carter BS, Bussemakers MJ, Emi M, Fujiwara Y, Kyprianou N et al. Homozygous deletion and frequent allelic loss of chromosome 8p22 loci in human prostate cancer. Cancer Res 1993; 53: 3869–3873.
Macoska JA, Trybus TM, Benson PD, Sakr WA, Grignon DJ, Wojno KD et al. Evidence for three tumor suppressor gene loci on chromosome 8p in human prostate cancer. Cancer Res 1995; 55: 5390–5395.
Kagan J, Stein J, Babaian RJ, Joe YS, Pisters LL, Glassman AB et al. Homozygous deletions at 8p22 and 8p21 in prostate cancer implicate these regions as the sites for candidate tumor suppressor genes. Oncogene 1995; 11: 2121–2126.
Lu W, Takahashi H, Furusato B, Maekawa S, Ikegami M, Sudo A et al. Allelotyping analysis at chromosome arm 8p of high-grade prostatic intraepithelial neoplasia and incidental, latent, and clinical prostate cancers. Genes Chromosomes Cancer 2006; 45: 509–515.
El Gammal AT, Bruchmann M, Zustin J, Isbarn H, Hellwinkel OJ, Kollermann J et al. Chromosome 8p deletions and 8q gains are associated with tumor progression and poor prognosis in prostate cancer. Clin Cancer Res 2010; 16: 56–64.
Calin GA, Sevignani C, Dumitru CD, Hyslop T, Noch E, Yendamuri S et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci USA 2004; 101: 2999–3004.
Bartel DP . MicroRNAs: target recognition and regulatory functions. Cell 2009; 136: 215–233.
Piovani G, Savio G, Traversa M, Pilotta A, De Petro G, Barlati S et al. De novo 1 Mb interstitial deletion of 8p22 in a patient with slight mental retardation and speech delay. Mol Cytogenet 2014; 7: 25.
Lian J, Tian H, Liu L, Zhang XS, Li WQ, Deng YM et al. Downregulation of microRNA-383 is associated with male infertility and promotes testicular embryonal carcinoma cell proliferation by targeting IRF1. Cell Death Dis 2010; 1: e94.
He Z, Cen D, Luo X, Li D, Li P, Liang L et al. Downregulation of miR-383 promotes glioma cell invasion by targeting insulin-like growth factor 1 receptor. Med Oncol 2013; 30: 557.
Huang H, Tian H, Duan Z, Cao Y, Zhang XS, Sun F . microRNA-383 impairs phosphorylation of H2AX by targeting PNUTS and inducing cell cycle arrest in testicular embryonal carcinoma cells. Cell Signal 2014; 26: 903–911.
Xu D, Ma P, Gao G, Gui Y, Niu X, Jin B . MicroRNA-383 expression regulates proliferation, migration, invasion, and apoptosis in human glioma cells. Tumour Biol 2015; 36: 7743–7753.
Xu Z, Zeng X, Tian D, Xu H, Cai Q, Wang J et al. MicroRNA-383 inhibits anchorage-independent growth and induces cell cycle arrest of glioma cells by targeting CCND1. Biochem Biophysi Res Commun 2014; 453: 833–838.
Zhao L, Gu H, Chang J, Wu J, Wang D, Chen S et al. MicroRNA-383 regulates the apoptosis of tumor cells through targeting Gadd45g. PLoS ONE 2014; 9: e110472.
Han S, Cao C, Tang T, Lu C, Xu J, Wang S et al. ROBO3 promotes growth and metastasis of pancreatic carcinoma. Cancer Lett 2015; 366: 61–70.
Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2012; 2: 401–404.
Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 2013; 6: pl1.
Betel D, Wilson M, Gabow A, Marks DS, Sander C . The microRNA.org resource: targets and expression. Nucleic Acids Res 2008; 36: D149–D153.
Abeshouse A, Ahn J, Akbani R, Ally A, Amin S, Andry CD et al. The molecular taxonomy of primary prostate cancer. Cell 2015; 163: 1011–1025.
Porkka KP, Pfeiffer MJ, Waltering KK, Vessella RL, Tammela TL, Visakorpi T . MicroRNA expression profiling in prostate cancer. Cancer Res 2007; 67: 6130–6135.
Matsuyama H, Pan Y, Yoshihiro S, Kudren D, Naito K, Bergerheim US et al. Clinical significance of chromosome 8p, 10q, and 16q deletions in prostate cancer. Prostate 2003; 54: 103–111.
Matsuyama H, Pan Y, Oba K, Yoshihiro S, Matsuda K, Hagarth L et al. The role of chromosome 8p22 deletion for predicting disease progression and pathological staging in prostate cancer. Aktuelle Urol 2003; 34: 247–249.
Matsuyama H, Oba K, Matsuda K, Yoshihiro S, Tsukamoto M, Kinjo M et al. Haploinsufficiency of 8p22 may influence cancer-specific survival in prostate cancer. Cancer Genet Cytogenet 2007; 174: 24–34.
Liu C, Kelnar K, Liu B, Chen X, Calhoun-Davis T, Li H et al. The microRNA miR-34a inhibits prostate cancer stem cells and metastasis by directly repressing CD44. Nat Med 2011; 17: 211–215.
Saini S, Majid S, Shahryari V, Arora S, Yamamura S, Chang I et al. miRNA-708 control of CD44(+) prostate cancer-initiating cells. Cancer Res 2012; 72: 3618–3630.
Yang K, Handorean AM, Iczkowski KA . MicroRNAs 373 and 520c are downregulated in prostate cancer, suppress CD44 translation and enhance invasion of prostate cancer cells in vitro. Int J Clin Exp Pathol 2009; 2: 361–369.
Krek A, Grun D, Poy MN, Wolf R, Rosenberg L, Epstein EJ et al. Combinatorial microRNA target predictions. Nat Genet 2005; 37: 495–500.
Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB . Prediction of mammalian microRNA targets. Cell 2003; 115: 787–798.
Liao XH, Zheng L, He HP, Zheng DL, Wei ZQ, Wang N et al. STAT3 regulated ATR via microRNA-383 to control DNA damage to affect apoptosis in A431 cells. Cell Signal 2015; 27: 2285–2295.
Hsieh IS, Chang KC, Tsai YT, Ke JY, Lu PJ, Lee KH et al. MicroRNA-320 suppresses the stem cell-like characteristics of prostate cancer cells by downregulating the Wnt/beta-catenin signaling pathway. Carcinogenesis 2013; 34: 530–538.
Yin M, Wang X, Yao G, Lu M, Liang M, Sun Y et al. Transactivation of micrornA-320 by microRNA-383 regulates granulosa cell functions by targeting E2F1 and SF-1 proteins. J Biol Chem 2014; 289: 18239–18257.
Brooks DD, Wolf A, Smith RA, Dash C, Guessous I . Prostate cancer screening 2010: updated recommendations from the American Cancer Society. J Natl Med Assoc 2010; 102: 423–429.
Wolf AM, Wender RC, Etzioni RB, Thompson IM, D'Amico AV, Volk RJ et al. American Cancer Society guideline for the early detection of prostate cancer: update 2010. CA Cancer J Clin 2010; 60: 70–98.
Saini S . PSA and beyond: alternative prostate cancer biomarkers. Cell Oncol (Dordr) 2016; 39: 97–106.
Saini S, Majid S, Yamamura S, Tabatabai L, Suh SO, Shahryari V et al. Regulatory Role of mir-203 in Prostate Cancer Progression and Metastasis. Clin Cancer Res 2011; 17: 5287–5298.
Acknowledgements
We thank Dr Roger Erickson for his support and assistance with preparation of the manuscript. We acknowledge UCSF CTSI (Dr Iryna V. Lobach) for help with statistical analysis. This work was supported by the National Cancer Institute at the National Institutes of Health (Grant Number RO1CA177984, RO1CA138642), VA program project on PCa (BX001604).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Oncogene website
Supplementary information
Rights and permissions
About this article
Cite this article
Bucay, N., Sekhon, K., Yang, T. et al. MicroRNA-383 located in frequently deleted chromosomal locus 8p22 regulates CD44 in prostate cancer. Oncogene 36, 2667–2679 (2017). https://doi.org/10.1038/onc.2016.419
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/onc.2016.419
This article is cited by
-
MiR-383 sensitizes osteosarcoma cells to bortezomib treatment via down-regulating PSMB5
Molecular Biology Reports (2024)
-
Exosomal miRNAs—a diagnostic biomarker acting as a guiding light in the diagnosis of prostate cancer
Functional & Integrative Genomics (2023)
-
MicroRNA-3613-3p functions as a tumor suppressor and represents a novel therapeutic target in breast cancer
Breast Cancer Research (2021)
-
Loss of RBMS1 as a regulatory target of miR-106b influences cell growth, gap closing and colony forming in prostate carcinoma
Scientific Reports (2020)
-
The biology and role of CD44 in cancer progression: therapeutic implications
Journal of Hematology & Oncology (2018)