Abstract
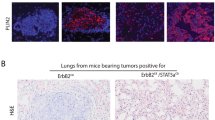
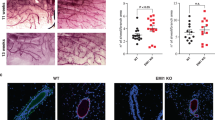
Metaplastic breast carcinoma is an aggressive form of invasive breast cancer with histological evidence of epithelial to mesenchymal transition (EMT). However, the defining molecular events are unknown. Here we show that CCN6 (WISP3), a secreted matricellular protein of the CCN (CYR61/CTGF/NOV) family, is significantly downregulated in clinical samples of human spindle cell metaplastic breast carcinoma. We generated a mouse model of mammary epithelial-specific Ccn6 deletion by developing a floxed Ccn6 mouse which was bred with an MMTV-Cre mouse. Ccn6fl/fl;MMTV-Cre mice displayed severe defects in ductal branching and abnormal age-related involution compared to littermate controls. Ccn6fl/fl;MMTV-Cre mice developed invasive high grade mammary carcinomas with bona fide EMT, histologically similar to human metaplastic breast carcinomas. Global gene expression profiling of Ccn6fl/fl mammary carcinomas and comparison of orthologous genes with a human metaplastic carcinoma signature revealed a significant overlap of 87 genes (P=5 × 10−11). Among the shared deregulated genes between mouse and human are important regulators of epithelial morphogenesis including Cdh1, Ck19, Cldn3 and 4, Ddr1, and Wnt10a. These results document a causal role for Ccn6 deletion in the pathogenesis of metaplastic carcinomas with histological and molecular similarities with human disease. We provide a platform to study new targets in the diagnosis and treatment of human metaplastic carcinomas, and a new disease relevant model in which to test new treatment strategies.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Oberman HA . Metaplastic carcinoma of the breast. A clinicopathologic study of 29 patients. Am J Surg Pathol 1987; 11: 918–929.
Bae SY, Lee SK, Koo MY, Hur SM, Choi MY, Cho DH et al. The prognoses of metaplastic breast cancer patients compared to those of triple-negative breast cancer patients. Breast Cancer Res Treat 2011; 126: 471–478.
Luini A, Aguilar M, Gatti G, Fasani R, Botteri E, Brito JA et al. Metaplastic carcinoma of the breast, an unusual disease with worse prognosis: the experience of the European Institute of Oncology and review of the literature. Breast Cancer Res Treat 2007; 101: 349–353.
Burstein MD, Tsimelzon A, Poage GM, Covington KR, Contreras A, Fuqua SA et al. Comprehensive genomic analysis identifies novel subtypes and targets of triple-negative breast cancer. Clin Cancer Res 2015; 21: 1688–1698.
Hennessy BT, Gonzalez-Angulo AM, Stemke-Hale K, Gilcrease MZ, Krishnamurthy S, Lee JS et al. Characterization of a naturally occurring breast cancer subset enriched in epithelial-to-mesenchymal transition and stem cell characteristics. Cancer Res 2009; 69: 4116–4124.
Weigelt B, Ng CK, Shen R, Popova T, Schizas M, Natrajan R et al. Metaplastic breast carcinomas display genomic and transcriptomic heterogeneity [corrected]. Mod Pathol 2015; 28: 340–351.
Hugo H, Ackland ML, Blick T, Lawrence MG, Clements JA, Williams ED et al. Epithelial-mesenchymal and mesenchymal-epithelial transitions in carcinoma progression. J Cell Physiol 2007; 213: 374–383.
Jun JI, Lau LF . Taking aim at the extracellular matrix: CCN proteins as emerging therapeutic targets. Nat Rev Drug Discov 2011; 10: 945–963.
Huang W, Zhang Y, Varambally S, Chinnaiyan AM, Banerjee M, Merajver SD et al. Inhibition of CCN6 (Wnt-1-induced signaling protein 3) down-regulates E-cadherin in the breast epithelium through induction of snail and ZEB1. Am J Pathol 2008; 172: 893–904.
Lau LF . CCN1/CYR61: the very model of a modern matricellular protein. Cell Mol Life Sci 2011; 68: 3149–3163.
Lorenzatti G, Huang W, Pal A, Cabanillas AM, Kleer CG . CCN6 (WISP3) decreases ZEB1-mediated EMT and invasion by attenuation of IGF-1 receptor signaling in breast cancer. J Cell Sci 2011; 124: 1752–1758.
Pal A, Huang W, Li X, Toy KA, Nikolovska-Coleska Z, Kleer CG . CCN6 modulates BMP signaling via the Smad-independent TAK1/p38 pathway, acting to suppress metastasis of breast cancer. Cancer Res 2012a; 72: 4818–4828.
Zhang Y, Pan Q, Zhong H, Merajver SD, Kleer CG. . Inhibition of CCN6 (WISP3) expression promotes neoplastic progression and enhances the effects of insulin-like growth factor-1 on breast epithelial cells. Breast Cancer Res 2005; 7: R1080–R1089.
Kleer CG, Zhang Y, Pan Q, Merajver SD . WISP3 (CCN6) is a secreted tumor-suppressor protein that modulates IGF signaling in inflammatory breast cancer. Neoplasia 2004; 6: 179–185.
van Golen KL, Davies S, Wu ZF, Wang YF, Bucana CD, Root H et al. A novel putative low-affinity insulin-like growth factor-binding protein, LIBC (lost in inflammatory breast cancer), and RhoC GTPase correlate with the inflammatory breast cancer phenotype. Clin Cancer Res 1999; 5: 2511–2519.
Huang W, Gonzalez ME, Toy KA, Banerjee M, Kleer CG . Blockade of CCN6 (WISP3) activates growth factor-independent survival and resistance to anoikis in human mammary epithelial cells. Cancer Res 2010; 70: 3340–3350.
Pal A, Huang W, Toy KA, Kleer CG . CCN6 knockdown disrupts acinar organization of breast cells in three-dimensional cultures through up-regulation of type III TGF-beta receptor. Neoplasia 2012b; 14: 1067–1074.
Milanese TR, Hartmann LC, Sellers TA, Frost MH, Vierkant RA, Maloney SD et al. Age-related lobular involution and risk of breast cancer. J Natl Cancer Inst 2006; 98: 1600–1607.
Radisky DC, Hartmann LC . Mammary involution and breast cancer risk: transgenic models and clinical studies. J Mammary Gland Biol Neoplasia 2009; 14: 181–191.
Guy CT, Cardiff RD, Muller WJ . Induction of mammary tumors by expression of polyomavirus middle T oncogene: a transgenic mouse model for metastatic disease. Mol Cell Biol 1992; 12: 954–961.
Koker MM, Kleer CG . p63 expression in breast cancer: a highly sensitive and specific marker of metaplastic carcinoma. Am J Surg Pathol 2004; 28: 1506–1512.
Rakha EA, Tan PH, Varga Z, Tse GM, Shaaban AM, Climent F et al. Prognostic factors in metaplastic carcinoma of the breast: a multi-institutional study. Br J Cancer 2015; 112: 283–289.
Reis-Filho JS, Milanezi F, Carvalho S, Simpson PT, Steele D, Savage K et al. Metaplastic breast carcinomas exhibit EGFR, but not HER2, gene amplification and overexpression: immunohistochemical and chromogenic in situ hybridization analysis. Breast Cancer Res 2005; 7: R1028–R1035.
Rungta S, Kleer CG . Metaplastic carcinomas of the breast: diagnostic challenges and new translational insights. Arch Pathol Lab Med 2012; 136: 896–900.
Reis-Filho JS, Milanezi F, Paredes J, Silva P, Pereira EM, Maeda SA et al. Novel and classic myoepithelial/stem cell markers in metaplastic carcinomas of the breast. Appl Immunohistochem Mol Morphol 2003; 11: 1–8.
Reis-Filho JS, Schmitt FC . p63 expression in sarcomatoid/metaplastic carcinomas of the breast. Histopathology 2003; 42: 94–95.
Hennessy BT, Giordano S, Broglio K, Duan Z, Trent J, Buchholz TA et al. Biphasic metaplastic sarcomatoid carcinoma of the breast. Ann Oncol 2006; 17: 605–613.
Hayes MJ, Thomas D, Emmons A, Giordano TJ, Kleer CG . Genetic changes of Wnt pathway genes are common events in metaplastic carcinomas of the breast. Clin Cancer Res 2008; 14: 4038–4044.
Anwar TE, Kleer CG . Tissue-based identification of stem cells and epithelial-to-mesenchymal transition in breast cancer. Hum Pathol 2013; 44: 1457–1464.
Hurvitz JR, Suwairi WM, Van Hul W, El-Shanti H, Superti-Furga A, Roudier J et al. Mutations in the CCN gene family member WISP3 cause progressive pseudorheumatoid dysplasia. Nat Genet 1999; 23: 94–98.
Pennica D, Swanson TA, Welsh JW, Roy MA, Lawrence DA, Lee J et al. WISP genes are members of the connective tissue growth factor family that are up-regulated in wnt-1-transformed cells and aberrantly expressed in human colon tumors. Proc Natl Acad Sci USA 1998; 95: 14717–14722.
Kutz WE, Gong Y, Warman ML . WISP3, the gene responsible for the human skeletal disease progressive pseudorheumatoid dysplasia, is not essential for skeletal function in mice. Mol Cell Biol 2005; 25: 414–421.
Huang W, Martin EE, Burman B, Gonzalez ME, Kleer CG . The matricellular protein ccn6 (wisp3) decreases notch1 and suppresses breast cancer initiating cells. Oncotarget 2016; 7: 25180–25193.
Kleer CG, Zhang Y, Pan Q, van Golen KL, Livant D et al. WISP3 is a novel tumor suppressor gene of inflammatory breast cancer. Oncogene 2002; 21: 3172–3180.
Ferguson BW, Gao X, Kil H, Lee J, Benavides F, Abba MC et al. Conditional Wwox deletion in mouse mammary gland by means of two Cre recombinase approaches. PLoS One 2012; 7: e36618.
McAllister KA, Bennett LM, Houle CD, Ward T, Malphurs J, Collins NK et al. Cancer susceptibility of mice with a homozygous deletion in the COOH-terminal domain of the Brca2 gene. Cancer Res 2002; 62: 990–994.
Xu X, Wagner KU, Larson D, Weaver Z, Li C, Ried T et al. Conditional mutation of Brca1 in mammary epithelial cells results in blunted ductal morphogenesis and tumour formation. Nat Genet 1999; 22: 37–43.
Mollenhauer J, Wiemann S, Scheurlen W, Korn B, Hayashi Y, Wilgenbus KK et al. DMBT1, a new member of the SRCR superfamily, on chromosome 10q25.3-26.1 is deleted in malignant brain tumours. Nat Genet 1997; 17: 32–39.
Blackburn AC, Hill LZ, Roberts AL, Wang J, Aud D, Jung J et al. Genetic mapping in mice identifies DMBT1 as a candidate modifier of mammary tumors and breast cancer risk. Am J Pathol 2007; 170: 2030–2041.
Tchatchou S, Riedel A, Lyer S, Schmutzhard J, Strobel-Freidekind O, Gronert-Sum S et al. Identification of a DMBT1 polymorphism associated with increased breast cancer risk and decreased promoter activity. Hum Mutat 2010; 31: 60–66.
Jones LP, Buelto D, Tago E, Owusu-Boaitey KE . Abnormal mammary adipose tissue environment of Brca1 mutant mice show a persistent deposition of highly vascularized multilocular adipocytes. J Cancer Sci Ther 2011; (Suppl 2)pii 004.
Zhang Y, Toy KA, Kleer CG . Metaplastic breast carcinomas are enriched in markers of tumor-initiating cells and epithelial to mesenchymal transition. Mod Pathol 2012; 25: 178–184.
Morishita A, Zaidi MR, Mitoro A, Sankarasharma D, Szabolcs M, Okada Y et al. HMGA2 is a driver of tumor metastasis. Cancer Res 2013; 73: 4289–4299.
Li Y, Francia G, Zhang JY . p62/IMP2 stimulates cell migration and reduces cell adhesion in breast cancer. Oncotarget 2015; 6: 32656–32668.
Zhou ZN, Sharma VP, Beaty BT, Roh-Johnson M, Peterson EA, Van Rooijen N et al. Autocrine HBEGF expression promotes breast cancer intravasation, metastasis and macrophage-independent invasion in vivo. Oncogene 2014; 33: 3784–3793.
Bernardo GM, Keri RA . FOXA1: a transcription factor with parallel functions in development and cancer. Biosci Rep 2012; 32: 113–130.
Parr C, Jiang WG . Hepatocyte growth factor activation inhibitors (HAI-1 and HAI-2) regulate HGF-induced invasion of human breast cancer cells. Int J Cancer 2006; 119: 1176–1183.
Toy KA, Valiathan RR, Nunez F, Kidwell KM, Gonzalez ME, Fridman R et al. Tyrosine kinase discoidin domain receptors DDR1 and DDR2 are coordinately deregulated in triple-negative breast cancer. Breast Cancer Res Treat 2015; 150: 9–18.
Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 2003; 4: 249–264.
Smyth GK . Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol 2004; 3: Article3.
Ritchie ME, Diyagama D, Neilson J, van Laar R, Dobrovic A, Holloway A et al. Empirical array quality weights in the analysis of microarray data. BMC Bioinform 2006; 7: 261.
Benjamini Y, Hochberg Y . Controlling the false discovery rate: a practical and powerful approach in multiple testing. J R Stat Soc 1995, Series B 57: 289–300.
Acknowledgements
This work was supported by the National Institutes of Health (NIH) grant R01CA125577 and R01CA107469 (C.G.K), F30CA196084 (T.A), R25GM086262 (PREP program, C.A-G), and the University of Michigan’s Cancer Center Support grant 5 P30 CA46592. We thank Dr Stephen Weiss for the MMTV-Cre line A mice in FVB background, and members of the Kleer lab for critical review of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Oncogene website
Supplementary information
Rights and permissions
About this article
Cite this article
Martin, E., Huang, W., Anwar, T. et al. MMTV-cre;Ccn6 knockout mice develop tumors recapitulating human metaplastic breast carcinomas. Oncogene 36, 2275–2285 (2017). https://doi.org/10.1038/onc.2016.381
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/onc.2016.381
This article is cited by
-
The matricellular protein CCN6 differentially regulates mitochondrial metabolism in normal epithelium and in metaplastic breast carcinomas
Journal of Cell Communication and Signaling (2022)
-
Mammary-specific expression of Trim24 establishes a mouse model of human metaplastic breast cancer
Nature Communications (2021)
-
A comprehensive overview of metaplastic breast cancer: clinical features and molecular aberrations
Breast Cancer Research (2020)
-
Quantitative proteomic landscape of metaplastic breast carcinoma pathological subtypes and their relationship to triple-negative tumors
Nature Communications (2020)
-
The emerging role of WISP proteins in tumorigenesis and cancer therapy
Journal of Translational Medicine (2019)