Abstract
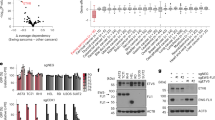
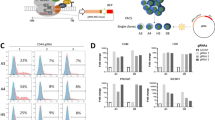
Ewing sarcoma is characterized by chromosomal translocations fusing the EWS gene with various members of the ETS family of transcription factors, most commonly FLI1. EWS-FLI1 is an aberrant transcription factor driving Ewing sarcoma tumorigenesis by either transcriptionally inducing or repressing specific target genes. Herein, we showed that Sprouty 1 (SPRY1), which is a physiological negative feedback inhibitor downstream of fibroblast growth factor (FGF) receptors (FGFRs) and other RAS-activating receptors, is an EWS-FLI1 repressed gene. EWS-FLI1 knockdown specifically increased the expression of SPRY1, while other Sprouty family members remained unaffected. Analysis of SPRY1 expression in a panel of Ewing sarcoma cells showed that SPRY1 was not expressed in Ewing sarcoma cell lines, suggesting that it could act as a tumor suppressor gene in these cells. In agreement, induction of SPRY1 in three different Ewing sarcoma cell lines functionally impaired proliferation, clonogenic growth and migration. In addition, SPRY1 expression inhibited extracellular signal-related kinase/mitogen-activated protein kinase (MAPK) signaling induced by serum and basic FGF (bFGF). Moreover, treatment of Ewing sarcoma cells with the potent FGFR inhibitor PD-173074 reduced bFGF-induced proliferation, colony formation and in vivo tumor growth in a dose-dependent manner, thus mimicking SPRY1 activity in Ewing sarcoma cells. Although the expression of SPRY1 was low when compared with other tumors, SPRY1 was variably expressed in primary Ewing sarcoma tumors and higher expression levels were significantly associated with improved outcome in a large patient cohort. Taken together, our data indicate that EWS-FLI1-mediated repression of SPRY1 leads to unrestrained bFGF-induced cell proliferation, suggesting that targeting the FGFR/MAPK pathway can constitute a promising therapeutic approach for this devastating disease.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Mackintosh C, Madoz-Gurpide J, Ordonez JL, Osuna D, Herrero-Martin D . The molecular pathogenesis of Ewing’s sarcoma. Cancer Biol Ther 2010; 9: 655–667.
Grohar PJ, Helman LJ . Prospects and challenges for the development of new therapies for Ewing sarcoma. Pharmacol Ther 2013; 137: 216–224.
Ladenstein R, Potschger U, Le Deley MC, Whelan J, Paulussen M, Oberlin O et al. Primary disseminated multifocal Ewing sarcoma: results of the Euro-EWING 99 trial. J Clin Oncol 2010; 28: 3284–3291.
Zhu L, McManus MM, Hughes DP . Understanding the biology of bone sarcoma from early initiating events through late events in metastasis and disease progression. Front Oncol 2013; 3: 230.
Delattre O, Zucman J, Plougastel B, Desmaze C, Melot T, Peter M et al. Gene fusion with an ETS DNA-binding domain caused by chromosome translocation in human tumours. Nature 1992; 359: 162–165.
Kovar H . Blocking the road, stopping the engine or killing the driver? Advances in targeting EWS/FLI-1 fusion in Ewing sarcoma as novel therapy. Expert Opin Ther Targets 2014; 18: 1315–1328.
Carrillo J, Garcia-Aragoncillo E, Azorin D, Agra N, Sastre A, Gonzalez-Mediero I et al. Cholecystokinin down-regulation by RNA interference impairs Ewing tumor growth. Clin Cancer Res 2007; 13: 2429–2440.
Garcia-Aragoncillo E, Carrillo J, Lalli E, Agra N, Gomez-Lopez G, Pestana A et al. DAX1, a direct target of EWS/FLI1 oncoprotein, is a principal regulator of cell-cycle progression in Ewing’s tumor cells. Oncogene 2008; 27: 6034–6043.
Smith R, Owen LA, Trem DJ, Wong JS, Whangbo JS, Golub TR et al. Expression profiling of EWS/FLI identifies NKX2.2 as a critical target gene in Ewing’s sarcoma. Cancer Cell 2006; 9: 405–416.
Surdez D, Benetkiewicz M, Perrin V, Han ZY, Pierron G, Ballet S et al. Targeting the EWSR1-FLI1 oncogene-induced protein kinase PKC-beta abolishes ewing sarcoma growth. Cancer Res 2012; 72: 4494–4503.
Grunewald TG, Diebold I, Esposito I, Plehm S, Hauer K, Thiel U et al. STEAP1 is associated with the invasive and oxidative stress phenotype of Ewing tumors. Mol Cancer Res 2012; 10: 52–65.
Prieur A, Tirode F, Cohen P, Delattre O . EWS/FLI-1 silencing and gene profiling of Ewing cells reveal downstream oncogenic pathways and a crucial role for repression of insulin-like growth factor binding protein 3. Mol Cell Biol 2004; 24: 7275–7283.
Agra N, Cidre F, Garcia-Garcia L, de la Parra J, Alonso J . Lysyl oxidase is downregulated by the EWS/FLI1 oncoprotein and its propeptide domain displays tumor supressor activities in ewing sarcoma cells. PLoS One 2013; 8: e66281.
Navarro D, Agra N, Pestana A, Alonso J, Gonzalez-Sancho JM . The EWS/FLI1 oncogenic protein inhibits expression of the Wnt inhibitor DICKKOPF-1 gene and antagonizes beta-catenin/TCF-mediated transcription. Carcinogenesis 2010; 31: 394–401.
Hahm KB, Cho K, Lee C, Im YH, Chang J, Choi SG et al. Repression of the gene encoding the TGF-beta type II receptor is a major target of the EWS-FLI1 oncoprotein. Nat Genet 1999; 23: 222–227.
Minowada G, Jarvis LA, Chi CL, Neubuser A, Sun X, Hacohen N et al. Vertebrate Sprouty genes are induced by FGF signaling and can cause chondrodysplasia when overexpressed. Development 1999; 126: 4465–4475.
Guy GR, Wong ES, Yusoff P, Chandramouli S, Lo TL, Lim J et al. Sprouty: how does the branch manager work? J Cell Sci 2003; 116: 3061–3068.
Christofori G . Split personalities: the agonistic antagonist Sprouty. Nat Cell Biol 2003; 5: 377–379.
Zhao Z, Zuber J, Diaz-Flores E, Lintault L, Kogan SC, Shannon K et al. p53 loss promotes acute myeloid leukemia by enabling aberrant self-renewal. Genes Dev 2010; 24: 1389–1402.
Fritzsche S, Kenzelmann M, Hoffmann MJ, Muller M, Engers R, Grone HJ et al. Concomitant down-regulation of SPRY1 and SPRY2 in prostate carcinoma. Endocr Relat Cancer 2006; 13: 839–849.
Lo TL, Yusoff P, Fong CW, Guo K, McCaw BJ, Phillips WA et al. The ras/mitogen-activated protein kinase pathway inhibitor and likely tumor suppressor proteins, sprouty 1 and sprouty 2 are deregulated in breast cancer. Cancer Res 2004; 64: 6127–6136.
Kwabi-Addo B, Ren C, Ittmann M . DNA methylation and aberrant expression of Sprouty1 in human prostate cancer. Epigenetics 2009; 4: 54–61.
Kwabi-Addo B, Wang J, Erdem H, Vaid A, Castro P, Ayala G et al. The expression of Sprouty1, an inhibitor of fibroblast growth factor signal transduction, is decreased in human prostate cancer. Cancer Res 2004; 64: 4728–4735.
Masoumi-Moghaddam S, Amini A, Ehteda A, Wei AQ, Morris DL . The expression of the Sprouty 1 protein inversely correlates with growth, proliferation, migration and invasion of ovarian cancer cells. J Ovarian Res 2014; 7: 61.
Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 2012; 483: 603–607.
Grunewald TG, Bernard V, Gilardi-Hebenstreit P, Raynal V, Surdez D, Aynaud MM et al. Chimeric EWSR1-FLI1 regulates the Ewing sarcoma susceptibility gene EGR2 via a GGAA microsatellite. Nat Genet 2015; 47: 1073–1078.
Willier S, Butt E, Grunewald TG . Lysophosphatidic acid (LPA) signalling in cell migration and cancer invasion: a focussed review and analysis of LPA receptor gene expression on the basis of more than 1700 cancer microarrays. Biol Cell 2013; 105: 317–333.
Postel-Vinay S, Veron AS, Tirode F, Pierron G, Reynaud S, Kovar H et al. Common variants near TARDBP and EGR2 are associated with susceptibility to Ewing sarcoma. Nat Genet 2012; 44: 323–327.
Cidre-Aranaz F, Alonso J . EWS/FLI1 target genes and therapeutic opportunities in Ewing sarcoma. Front Oncol 2015; 5: 162.
Bilke S, Schwentner R, Yang F, Kauer M, Jug G, Walker RL et al. Oncogenic ETS fusions deregulate E2F3 target genes in Ewing sarcoma and prostate cancer. Genome Res 2013; 23: 1797–1809.
Riggi N, Knoechel B, Gillespie SM, Rheinbay E, Boulay G, Suva ML et al. EWS-FLI1 utilizes divergent chromatin remodeling mechanisms to directly activate or repress enhancer elements in Ewing sarcoma. Cancer Cell 2014; 26: 668–681.
Tomazou EM, Sheffield NC, Schmidl C, Schuster M, Schonegger A, Datlinger P et al. Epigenome mapping reveals distinct modes of gene regulation and widespread enhancer reprogramming by the oncogenic fusion protein EWS-FLI1. Cell Rep 2015; 10: 1082–1095.
Calvisi DF, Ladu S, Gorden A, Farina M, Lee JS, Conner EA et al. Mechanistic and prognostic significance of aberrant methylation in the molecular pathogenesis of human hepatocellular carcinoma. J Clin Invest 2007; 117: 2713–2722.
Macia A, Gallel P, Vaquero M, Gou-Fabregas M, Santacana M, Maliszewska A et al. Sprouty1 is a candidate tumor-suppressor gene in medullary thyroid carcinoma. Oncogene 2012; 31: 3961–3972.
Gross I, Bassit B, Benezra M, Licht JD . Mammalian sprouty proteins inhibit cell growth and differentiation by preventing ras activation. J Biol Chem 2001; 276: 46460–46468.
Mekkawy AH, Pourgholami MH, Morris DL . Human Sprouty1 suppresses growth, migration, and invasion in human breast cancer cells. Tumour Biol 2014; 35: 5037–5048.
Wiles ET, Lui-Sargent B, Bell R, Lessnick SL . BCL11B is up-regulated by EWS/FLI and contributes to the transformed phenotype in Ewing sarcoma. PLoS ONE 2013; 8: e59369.
Liu X, Lan Y, Zhang D, Wang K, Wang Y, Hua ZC . SPRY1 promotes the degradation of uPAR and inhibits uPAR-mediated cell adhesion and proliferation. Am J Cancer Res 2014; 4: 683–697.
Powers CJ, McLeskey SW, Wellstein A . Fibroblast growth factors, their receptors and signaling. Endocr Relat Cancer 2000; 7: 165–197.
Bottcher RT, Niehrs C . Fibroblast growth factor signaling during early vertebrate development. Endocr Rev 2005; 26: 63–77.
Chalkiadaki G, Nikitovic D, Berdiaki A, Sifaki M, Krasagakis K, Katonis P et al. Fibroblast growth factor-2 modulates melanoma adhesion and migration through a syndecan-4-dependent mechanism. Int J Biochem Cell Biol 2009; 41: 1323–1331.
Yamaguchi F, Saya H, Bruner JM, Morrison RS . Differential expression of two fibroblast growth factor-receptor genes is associated with malignant progression in human astrocytomas. Proc Natl Acad Sci USA 1994; 91: 484–488.
Touat M, Ileana E, Postel-Vinay S, Andre F, Soria JC . Targeting FGFR signaling in cancer. Clin Cancer Res 2015; 21: 2684–2694.
Kamura S, Matsumoto Y, Fukushi JI, Fujiwara T, Iida K, Okada Y et al. Basic fibroblast growth factor in the bone microenvironment enhances cell motility and invasion of Ewing’s sarcoma family of tumours by activating the FGFR1-PI3K-Rac1 pathway. Br J Cancer 2010; 103: 370–381.
Agelopoulos K, Richter GH, Schmidt E, Dirksen U, von Heyking K, Moser B et al. Deep sequencing in conjunction with expression and functional analyses reveals activation of FGFR1 in Ewing sarcoma. Clin Cancer Res 2015; 21: 4935–4946.
Tirode F, Surdez D, Ma X, Parker M, Le Deley MC, Bahrami A et al. Genomic landscape of Ewing sarcoma defines an aggressive subtype with co-association of STAG2 and TP53 mutations. Cancer Discov 2014; 4: 1342–1353.
Kovar H, Jug G, Aryee DN, Zoubek A, Ambros P, Gruber B et al. Among genes involved in the RB dependent cell cycle regulatory cascade, the p16 tumor suppressor gene is frequently lost in the Ewing family of tumors. Oncogene 1997; 15: 2225–2232.
Kovar H, Pospisilova S, Jug G, Printz D, Gadner H . Response of Ewing tumor cells to forced and activated p53 expression. Oncogene 2003; 22: 3193–3204.
Gaspar N, Hawkins DS, Dirksen U, Lewis IJ, Ferrari S, Le Deley MC et al. Ewing sarcoma: current management and future approaches through collaboration. J Clin Oncol 2015; 33: 3036–3046.
Terada N, Shiraishi T, Zeng Y, Aw-Yong KM, Liu Z, Takahashi S et al. Correlation of Sprouty1 and Jagged1 with aggressive prostate cancer cells with different sensitivities to androgen deprivation. J Cell Biochem 2014; 115: 1505–1515.
Mendiola M, Carrillo J, Garcia E, Lalli E, Hernandez T, de Alava E et al. The orphan nuclear receptor DAX1 is up-regulated by the EWS/FLI1 oncoprotein and is highly expressed in Ewing tumors. Int J Cancer 2006; 118: 1381–1389.
Yue PY, Leung EP, Mak NK, Wong RN . A simplified method for quantifying cell migration/wound healing in 96-well plates. J Biomol Screen 2010; 15: 427–433.
Acknowledgements
FC-A, LG-G, JCL, AS, PG-M, SEL-P, SM and JA are supported by Asociación Pablo Ugarte and Miguelañez SA, ASION-La Hucha de Tomás, Fundación La Sonrisa de Alex and Instituto de Salud Carlos III (PI12/00816 and Spanish Cancer Network RTICC RD12/0036/0027). TGPG is supported by a grant from ‘Verein zur Förderung von Wissenschaft und Forschung an der Medizinischen Fakultät der LMU München (WiFoMed)’, the Daimler and Benz Foundation in cooperation with the Reinhard Frank Foundation, by LMU Munich’s Institutional Strategy LMUexcellent within the framework of the German Excellence Initiative, the ‘Mehr LEBEN für krebskranke Kinder—Bettina-Bräu-Stiftung’, the Walter Schulz Foundation, the Fritz Thyssen Foundation (FTH-40.15.0.030MN) and by the German Cancer Aid (DKH-111886 and DKH-70112257). The ‘Genetics and Biology of Cancers’ team (TGPG, DS and OD) is supported by grants from the Ligue Nationale Contre Le Cancer (Equipe labellisée). This work was also supported by the European PROVABES, ASSET and EEC FP7 grants. We also thank the following associations for their invaluable support: the Société Française des Cancers de l’Enfant, Courir pour Mathieu, Dans les pas du Géant, Olivier Chape, Les Bagouzamanon, Enfants et Santé and les Amis de Claire. We thank Dr S Navarro (University of Valencia, Valencia, Spain) and Dr TJ Triche (Children’s Hospital Los Angeles, Los Angeles, USA) for providing us with Ewing sarcoma cell lines A4573 and TTC-466, respectively.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Oncogene website
Supplementary information
Rights and permissions
About this article
Cite this article
Cidre-Aranaz, F., Grünewald, T., Surdez, D. et al. EWS-FLI1-mediated suppression of the RAS-antagonist Sprouty 1 (SPRY1) confers aggressiveness to Ewing sarcoma. Oncogene 36, 766–776 (2017). https://doi.org/10.1038/onc.2016.244
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/onc.2016.244
This article is cited by
-
Integrated analysis reveals FLI1 regulates the tumor immune microenvironment via its cell-type-specific expression and transcriptional regulation of distinct target genes of immune cells in breast cancer
BMC Genomics (2024)
-
Targeting of AKT-Signaling Pathway Potentiates the Anti-cancer Efficacy of Doxorubicin in A673 Ewing Sarcoma Cell Line
BioNanoScience (2021)
-
Loss of Spry1 reduces growth of BRAFV600-mutant cutaneous melanoma and improves response to targeted therapy
Cell Death & Disease (2020)
-
Targeting the undruggable: exploiting neomorphic features of fusion oncoproteins in childhood sarcomas for innovative therapies
Cancer and Metastasis Reviews (2019)
-
Ewing sarcoma
Nature Reviews Disease Primers (2018)