Abstract
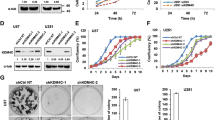
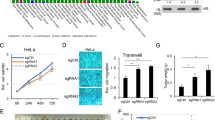
It is generally known that histone demethylases regulate gene transcription by altering the methylate status on histones, but their roles in cancers and the underlying molecular mechanisms still remain unclear. MYC-induced nuclear antigen (MINA) is reported to be a histone demethylase and highly expressed in many cancers. Here, for the first time, we show that MINA is involved in glioblastoma carcinogenesis and reveal the probable mechanisms of it in cell-cycle control. Kaplan–Meier analysis of progression-free survival showed that high MINA expression was strongly correlated with poor outcome and advancing tumor stage. MINA knockdown significantly repressed the cell proliferation and tumorigenesis abilities of glioblastoma cells in vitro and in vivo that were rescued by overexpressing the full-length MINA afterwards. Microarray analysis after knockdown of MINA revealed that MINA probably regulated glioblastoma carcinogenesis through the predominant cell-cycle pathways. Further investigation showed that MINA deficiency led to a cell-cycle arrest in G1 and G2 phases. And among the downstream genes, we found that cyclins and cyclin-dependent kinases were directly activated by MINA via the demethylation of H3K9me3.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Stupp R, Mason WP, van den Bent MJ, Weller M, Fisher B, Taphoorn MJ et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med 2005; 352: 987–996.
Benjamin R, Capparella J, Brown A . Classification of glioblastoma multiforme in adults by molecular genetics. Cancer J 2003; 9: 82–90.
Weller M, Cloughesy T, Perry JR, Wick W . Standards of care for treatment of recurrent glioblastoma—are we there yet? Neuro Oncol 2013; 15: 4–27.
Xie Q, Mittal S, Berens ME . Targeting adaptive glioblastoma: an overview of proliferation and invasion. Neuro Oncol 2014; 16: 1575–1584.
Tsuneoka M, Koda Y, Soejima M, Teye K, Kimura H . A novel myc target gene, mina53, that is involved in cell proliferation. J Biol Chem 2002; 277: 35450–35459.
Zhang Y, Lu Y, Yuan BZ, Castranova V, Shi X, Stauffer JL et al. The Human mineral dust-induced gene, mdig, is a cell growth regulating gene associated with lung cancer. Oncogene 2005; 24: 4873–4882.
Eilbracht J, Kneissel S, Hofmann A, Schmidt-Zachmann MS . Protein NO52—a constitutive nucleolar component sharing high sequence homologies to protein NO66. Eur J Cell Biol 2005; 84: 279–294.
Tan XP, Dong WG, Zhang Q, Yang ZR, Lei XF, Ai MH . Potential effects of Mina53 on tumor growth in human pancreatic cancer. Cell Biochem Biophys 2014; 69: 619–625.
Xing J, Wang K, Liu PW, Miao Q, Chen XY . Mina53, a novel molecular marker for the diagnosis and prognosis of gastric adenocarcinoma. Oncol Rep 2014; 31: 634–640.
Lu Y, Chang Q, Zhang Y, Beezhold K, Rojanasakul Y, Zhao H et al. Lung cancer-associated JmjC domain protein mdig suppresses formation of tri-methyl lysine 9 of histone H3. Cell Cycle 2009; 8: 2101–2109.
Komiya K, Sueoka-Aragane N, Sato A, Hisatomi T, Sakuragi T, Mitsuoka M et al. Expression of Mina53, a novel c-Myc target gene, is a favorable prognostic marker in early stage lung cancer. Lung Cancer 2010; 69: 232–238.
Teye K, Arima N, Nakamura Y, Sakamoto K, Sueoka E, Kimura H et al. Expression of Myc target gene mina53 in subtypes of human lymphoma. Oncol Rep 2007; 18: 841–848.
Teye K, Tsuneoka M, Arima N, Koda Y, Nakamura Y, Ueta Y et al. Increased expression of a Myc target gene Mina53 in human colon cancer. Am J Pathol 2004; 164: 205–216.
Tsuneoka M, Fujita H, Arima N, Teye K, Okamura T, Inutsuka H et al. Mina53 as a potential prognostic factor for esophageal squamous cell carcinoma. Clin Cancer Res 2004; 10: 7347–7356.
Fukahori S, Yano H, Tsuneoka M, Tanaka Y, Yagi M, Kuwano M et al. Immunohistochemical expressions of Cap43 and Mina53 proteins in neuroblastoma. J Pediatr Surg 2007; 42: 1831–1840.
Tan XP, Zhang Q, Dong WG, Lei XW, Yang ZR . Upregulated expression of Mina53 in cholangiocarcinoma and its clinical significance. Oncol Lett 2012; 3: 1037–1041.
Yu M, Sun J, Thakur C, Chen B, Lu Y, Zhao H et al. Paradoxical roles of mineral dust induced gene on cell proliferation and migration/invasion. PLoS One 2014; 9: e87998.
Lin H, Wang Y, Wang Y, Tian F, Pu P, Yu Y et al. Coordinated regulation of active and repressive histone methylations by a dual-specificity histone demethylase ceKDM7A from Caenorhabditis elegans. Cell Res 2010; 20: 899–907.
Chowdhury R, Sekirnik R, Brissett NC, Krojer T, Ho CH, Ng SS et al. Ribosomal oxygenases are structurally conserved from prokaryotes to humans. Nature 2014; 510: 422–426.
Chen B, Yu M, Chang Q, Lu Y, Thakur C, Ma D et al. Mdig de-represses H19 large intergenic non-coding RNA (lincRNA) by down-regulating H3K9me3 and heterochromatin. Oncotarget 2013; 4: 1427–1437.
Cohen I, Poreba E, Kamieniarz K, Schneider R . Histone modifiers in cancer: friends or foes? Genes Cancer 2011; 2: 631–647.
Lim S, Kaldis P . Cdks, cyclins and CKIs: roles beyond cell cycle regulation. Development 2013; 140: 3079–3093.
Kouzarides T . Chromatin modifications and their function. Cell 2007; 128: 693–705.
Jenuwein T, Allis CD . Translating the histone code. Science 2001; 293: 1074–1080.
Li B, Carey M, Workman JL . The role of chromatin during transcription. Cell 2007; 128: 707–719.
Shilatifard A . Chromatin modifications by methylation and ubiquitination: implications in the regulation of gene expression. Annu Rev Biochem 2006; 75: 243–269.
Kim TD, Shin S, Berry WL, Oh S, Janknecht R . The JMJD2A demethylase regulates apoptosis and proliferation in colon cancer cells. J Cell Biochem 2012; 113: 1368–1376.
Toyokawa G, Cho HS, Iwai Y, Yoshimatsu M, Takawa M, Hayami S et al. The histone demethylase JMJD2B plays an essential role in human carcinogenesis through positive regulation of cyclin-dependent kinase 6. Cancer Prev Res (Phila) 2011; 4: 2051–2061.
Zhao E, Ding J, Xia Y, Liu M, Ye B, Choi JH et al. KDM4C and ATF4 cooperate in transcriptional control of amino acid metabolism. Cell Rep 2016; 14: 506–519.
Komiya K, Sueoka-Aragane N, Sato A, Hisatomi T, Sakuragi T, Mitsuoka M et al. Mina53, a novel c-Myc target gene, is frequently expressed in lung cancers and exerts oncogenic property in NIH/3T3 cells. J Cancer Res Clin Oncol 2010; 136: 465–473.
Wang M, Liu Y, Zou J, Yang R, Xuan F, Wang Y et al. Transcriptional co-activator TAZ sustains proliferation and tumorigenicity of neuroblastoma by targeting CTGF and PDGF-β. Oncotarget 2015; 6: 9517–9530.
Yang R, Wu Y, Wang M, Sun Z, Zou J, Zhang Y et al. HDAC9 promotes glioblastoma growth via TAZ-mediated EGFR pathway activation. Oncotarget 2015; 6: 7644–7656.
Xuan F, Huang M, Liu W, Ding H, Yang L, Cui H . Homeobox C9 suppresses Beclin1-mediated autophagy in glioblastoma by directly inhibiting the transcription of death-associated protein kinase 1. Neuro Oncol 2015; 18: 819–829.
Acknowledgements
This work was supported by the National Basic Research Program of China (No. 2012CB114603), the National Natural Science Foundation of China (No. 31501100, 81502574), the Research Fund for the Doctoral Program of Higher Education of China (20130182110003) and the Basal Research Fund of Central Higher Education Institutions (XDJK2016D003).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Oncogene website
Rights and permissions
About this article
Cite this article
Huang, MY., Xuan, F., Liu, W. et al. MINA controls proliferation and tumorigenesis of glioblastoma by epigenetically regulating cyclins and CDKs via H3K9me3 demethylation. Oncogene 36, 387–396 (2017). https://doi.org/10.1038/onc.2016.208
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/onc.2016.208
This article is cited by
-
MESP2 binds competitively to TCF4 to suppress gastric cancer progression by regulating the SKP2/p27 axis
Cell Death Discovery (2023)
-
MDIG-mediated H3K9me3 demethylation upregulates Myc by activating OTX2 and facilitates liver regeneration
Signal Transduction and Targeted Therapy (2023)
-
The role of histone H3 lysine demethylases in glioblastoma
Cancer and Metastasis Reviews (2023)
-
JMJD family proteins in cancer and inflammation
Signal Transduction and Targeted Therapy (2022)
-
CSN6 promotes melanoma proliferation and metastasis by controlling the UBR5-mediated ubiquitination and degradation of CDK9
Cell Death & Disease (2021)