Abstract
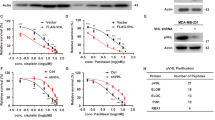
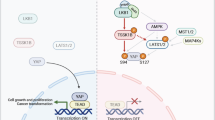
P21 Activated Kinase 1 (Pak1), an oncogenic serine/threonine kinase, is known to have a significant role in the regulation of cytoskeleton and cellular morphology. Runx3 was initially known for its role in tumor suppressor function, but recent studies have reported the oncogenic role of Runx3 in various cancers. However, the mechanism that controls the paradoxical functions of Runx3 still remains unclear. In this study, we show that Runx3 is a physiologically interacting substrate of Pak1. We identified the site of phosphorylation in Runx3 as Threonine 209 by mass spectrometry analysis and site-directed mutagenesis, and further confirmed the same with a site-specific antibody. Results from our functional studies showed that Threonine 209 phosphorylation in Runx3 alters its subcellular localization by protein mislocalization from the nucleus to the cytoplasm and subsequently converses its biological functions. This was further supported by in vivo tumor xenograft studies in nude mouse models which clearly demonstrated that PANC-28 cells transfected with the Runx3-T209E clone showed high tumorigenic potential as compared with other clones. Our results from clinical samples also suggest that Threonine 209 phosphorylation by Pak1 could be a potential therapeutic target and of great clinical relevance with implications for Runx3 inactivation in cancer cells where Runx3 is known to be oncogenic. The findings presented in this study provide evidence of Runx3-Threonine 209 phosphorylation as a molecular switch in dictating the tissue-specific dualistic functions of Runx3 for the first time.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Chuang LS, Ito K, Ito Y . RUNX family: Regulation and diversification of roles through interacting proteins. Int J Cancer 2013; 132: 1260–1271.
Kudo Y, Tsunematsu T, Takata T . Oncogenic role of RUNX3 in head and neck cancer. J Cell Biochem 2011; 112: 387–393.
Chuang LS, Ito Y . RUNX3 is multifunctional in carcinogenesis of multiple solid tumors. Oncogene 2010; 29: 2605–2615.
Bae SC, Choi JK . Tumor suppressor activity of RUNX3. Oncogene 2004; 23: 4336–4340.
Ito K, Liu Q, Salto-Tellez M, Yano T, Tada K, Ida H et al. RUNX3, A novel tumor suppressor, is frequently inactivated in gastric cancer by protein mislocalization. Cancer Res 2005; 65: 7743–7750.
Li QL, Ito K, Sakakura C, Fukamachi H, Ki I, Chi XZ et al. Causal relationship between the loss of RUNX3 expression and gastric cancer. Cell 2002; 109: 113–124.
Yanagawa N, Tamura G, Oizumi H, Takahashi N, Shimazaki Y, Motoyama T . Promoter hypermethylation of tumor suppressor and tumor-related genes in non-small cell lung cancers. Cancer Sci 2003; 94: 589–592.
Kim WJ, Kim EJ, Jeong P, Quan C, Kim J, Li QL et al. RUNX3 inactivation by point mutations and aberrant DNA methylation in bladder tumors. Cancer Res 2005; 65: 9347–9354.
Lau QC, Raja E, Salto-Tellez M, Liu Q, Ito K, Inoue M et al. RUNX3 is frequently inactivated by dual mechanisms of protein mislocalization and promoter hypermethylation in breast cancer. Cancer Res 2006; 66: 6512–6520.
Bae SC, Lee YH . Phosphorylation, acetylation and ubiquitination: the molecular basis of RUNX regulation. Gene 2006; 366: 58–66.
Goh YM, Cinghu S, Hong ET, Lee YS, Kim JH, Jang JW et al. Src kinase phosphorylates RUNX3 at tyrosine residues and localizes the protein in the cytoplasm. J BiolChem 2010; 285: 10122–10129.
Nicole Tsang YH, Wu XW, Lim JS, Wee Ong C, Salto-Tellez M, Ito K et al. Prolyl isomerase Pin1 downregulates tumor suppressor RUNX3 in breast cancer. Oncogene 2012; 32: 1488–1496.
Kumar R, Gururaj AE, Barnes CJ . p21-activated kinases in cancer. Nat Rev Cancer 2006; 6: 459–471.
Chi XZ, Yang JO, Lee KY, Ito K, Sakakura C, Li QL et al. RUNX3 suppresses gastric epithelial cell growth by inducing p21WAF1/Cip1 expression in cooperation with transforming growth factor β-activated SMAD. Mol Cell Biol 2005; 25: 8097–8107.
Lee CW, Chuang LS, Kimura S, Lai SK, Ong CW, Yan B et al. RUNX3 functions as an oncogene in ovarian cancer. Gynecol Oncol 2011; 122: 410–417.
Yoshiaki Ito . Oncogenic potential of the RUNX gene family: 'Overview'. Oncogene 2004; 23: 4198–4208.
Martin CW, Kamel I, Rani PG, Libing F, Markus AC, Kathleen ED et al. RUNX3 controls a metastatic switch in pancreatic ductal adenocarcinoma. Cell 2015; 161: 1–16.
Tsunematsu T, Kudo Y, Iizuka S, Ogawa I, Fujita T, Kurihara H et al. Runx3 has an oncogenic role in Head and Neck Cancer. PLoS One 2009; 4: e5892.
Chauang LS, Ito K, Ito Y . RUNX family: Regulation and diversification of roles through interacting proteins. Int J Cancer 2013; 15: 1260–1271.
Acknowledgements
We thank Prof Yoshiaki Ito, Cancer Science Institute of Singapore, Singapore, for providing us the Runx3 constructs. We profusely thank Prof Susanne M Gollin, University of Pittsburgh, for providing us with SCC-131 cells. We thank Dr Marsha Fraizer (University of Texas, M. D. Anderson Cancer Center, USA); for providing MDA Panc 28 cells. We thank Dr Rajeshwari, Bioklone, Chennai, for generating polyclonal Runx3-Threonine 209 phospho antibody. We thank DAE-BRNS (Grant # 35/14/41/2014-BRNS-RTAC), DBT (BT/PR14340/NNT/28/860/2015) Government of India for financial support to SKR and the Department of Biotechnology, Indian Institute of Technology Madras (IITM) for infrastructural facilities. We thank Mr Balabhaskara Rao and Ms Lakshmi Arivazhagan for the initial help with protein expression studies and Runx3 clones. We thank Professor SP Thyagarajan, Dean Research and Management of SRU for Support and encouragement.
Author contributions
AKC, MS, CC, SSS, RS, ST, RK and SJ – performed experiments; SS and KR – stained and scored the IHC slides; SA and DB – helped with immunofluorescence studies; RSP – performed statistical analysis; ASN – helped with clinical samples; GV, MS and SKR – designed the study and experiments and wrote the paper.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Oncogene website
Rights and permissions
About this article
Cite this article
Kumar, A., Singhal, M., Chopra, C. et al. Threonine 209 phosphorylation on RUNX3 by Pak1 is a molecular switch for its dualistic functions. Oncogene 35, 4857–4865 (2016). https://doi.org/10.1038/onc.2016.18
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/onc.2016.18
This article is cited by
-
EZH2 regulates a SETDB1/ΔNp63α axis via RUNX3 to drive a cancer stem cell phenotype in squamous cell carcinoma
Oncogene (2022)
-
Small peptide inhibitor from the sequence of RUNX3 disrupts PAK1–RUNX3 interaction and abrogates its phosphorylation-dependent oncogenic function
Oncogene (2021)
-
Elevated expression of RUNX3 co-expressing with EZH2 in esophageal cancer patients from India
Cancer Cell International (2020)