Abstract
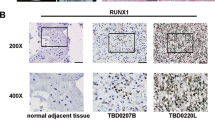
An integrative functional genomics study of multiple forms of data are vital for discovering molecular drivers of cancer development and progression. Here, we present an integrated genomic strategy utilizing DNA methylation and transcriptome profile data to discover epigenetically regulated genes implicated in cancer development and invasive progression. More specifically, this analysis identified fibromodulin (FMOD) as a glioblastoma (GBM) upregulated gene because of the loss of promoter methylation. Secreted FMOD promotes glioma cell migration through its ability to induce filamentous actin stress fiber formation. Treatment with cytochalasin D, an actin polymerization inhibitor, significantly reduced the FMOD-induced glioma cell migration. Small interfering RNA and small molecule inhibitor-based studies identified that FMOD-induced glioma cell migration is dependent on integrin-FAK-Src-Rho-ROCK signaling pathway. FMOD lacking C-terminus LRR11 domain (ΔFMOD), which does not bind collagen type I, failed to induce integrin and promote glioma cell migration. Further, FMOD-induced integrin activation and migration was abrogated by a 9-mer wild-type peptide from the FMOD C-terminus. However, the same peptide with mutation in two residues essential for FMOD interaction with collagen type I failed to compete with FMOD, thus signifying the importance of collagen type I–FMOD interaction in integrin activation. Chromatin immunoprecipitation–PCR experiments revealed that transforming growth factor beta-1 (TGF-β1) regulates FMOD expression through epigenetic remodeling of FMOD promoter that involved demethylation and gain of active histone marks with a simultaneous loss of DNMT3A and EZH2 occupancy, but enrichment of Sma- and Mad-related protein-2 (SMAD2) and CBP. FMOD silencing inhibited the TGF-β1-mediated glioma cell migration significantly. In univariate and multivariate Cox regression analysis, both FMOD promoter methylation and transcript levels predicted prognosis in GBM. Thus, this study identified several epigenetically regulated alterations responsible for cancer development and progression. Specifically, we found that secreted FMOD as an important regulator of glioma cell migration downstream of TGF-β1 pathway and forms a potential basis for therapeutic intervention in GBM.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Abbreviations
- ChIP:
-
chromatin immunoprecipitation
- FAK:
-
focal adhesion kinase
- FMOD:
-
fibromodulin
- GBM:
-
glioblastoma
- ROCK:
-
Rho-associated coiled-coil containing protein Kinase 1
- SMADs:
-
Sma- and Mad-related proteins
- TGF-β1:
-
transforming growth factor beta-1.
References
Cancer Genome Atlas Research N. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature 2008; 455: 1061–1068.
Verhaak RG, Hoadley KA, Purdom E, Wang V, Qi Y, Wilkerson MD et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell 2010; 17: 98–110.
Noushmehr H, Weisenberger DJ, Diefes K, Phillips HS, Pujara K, Berman BP et al. Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell 2010; 17: 510–522.
Frattini V, Trifonov V, Chan JM, Castano A, Lia M, Abate F et al. The integrated landscape of driver genomic alterations in glioblastoma. Nat Genet 45: 1141–1149.
Brennan CW, Verhaak RG, McKenna A, Campos B, Noushmehr H, Salama SR et al. The somatic genomic landscape of glioblastoma. Cell 2013; 155: 462–477.
Colman H, Zhang L, Sulman EP, McDonald JM, Shooshtari NL, Rivera A et al. A multigene predictor of outcome in glioblastoma. Neuro Oncol 2010; 12: 49–57.
Hegi ME, Diserens AC, Gorlia T, Hamou MF, de Tribolet N, Weller M et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med 2005; 352: 997–1003.
Parsons DW, Jones S, Zhang X, Lin JC, Leary RJ, Angenendt P et al. An integrated genomic analysis of human glioblastoma multiforme. Science 2008; 321: 1807–1812.
Shukla S, Bhargawa S, Somasundaram K . Cancer gene signatures in risk stratification. Current Science 2014; 107: 815–823.
Srinivasan S, Patric IR, Somasundaram K . A ten-microRNA expression signature predicts survival in glioblastoma. PLoS One 2011; 6: e17438.
Stupp R, Hegi ME, Mason WP, van den Bent MJ, Taphoorn MJ, Janzer RC et al. Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol 2009; 10: 459–466.
Chinot OL, Wick W, Mason W, Henriksson R, Saran F, Nishikawa R et al. Bevacizumab plus radiotherapy-temozolomide for newly diagnosed glioblastoma. N Engl J Med 2014; 370: 709–722.
Nathanson DA, Gini B, Mottahedeh J, Visnyei K, Koga T, Gomez G et al. Targeted therapy resistance mediated by dynamic regulation of extrachromosomal mutant EGFR DNA. Science 2014; 343: 72–76.
Meyer M, Reimand J, Lan X, Head R, Zhu X, Kushida M et al. Single cell-derived clonal analysis of human glioblastoma links functional and genomic heterogeneity. Proc Natl Acad Sci USA 2015; 112: 851–856.
Patel AP, Tirosh I, Trombetta JJ, Shalek AK, Gillespie SM, Wakimoto H et al. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science 2014; 344: 1396–1401.
Holland EC . Glioblastoma multiforme: the terminator. Proc Natl Acad Sci USA 2000; 97: 6242–6244.
Scherer HJ . A critical review: the pathology of cerebral gliomas. J Neurol Psychiatry 1940; 3: 147–177.
Beiko J, Suki D, Hess KR, Fox BD, Cheung V, Cabral M et al. IDH1 mutant malignant astrocytomas are more amenable to surgical resection and have a survival benefit associated with maximal surgical resection. Neuro Oncol 2014; 16: 81–91.
Shukla S, Pia Patric IR, Thinagararjan S, Srinivasan S, Mondal B, Hegde AS et al. A DNA methylation prognostic signature of glioblastoma: identification of NPTX2-PTEN-NF-kappaB nexus. Cancer Res 2013; 73: 6563–6573.
Huang, da W, Sherman BT, Lempicki RA . Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 2009; 4: 44–57.
Huang, da W, Sherman BT, Lempicki RA . Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 2009; 37: 1–13.
Vega FM, Ridley AJ . SnapShot: Rho family GTPases. Cell 2007; 129: 1430.
Ridley AJ, Hall A . The small GTP-binding protein rho regulates the assembly of focal adhesions and actin stress fibers in response to growth factors. Cell 1992; 70: 389–399.
Mouneimne G, Hansen SD, Selfors LM, Petrak L, Hickey MM, Gallegos LL et al. Differential remodeling of actin cytoskeleton architecture by profilin isoforms leads to distinct effects on cell migration and invasion. Cancer Cell 2012; 22: 615–630.
Stricker J, Falzone T, Gardel ML . Mechanics of the F-actin cytoskeleton. J Biomech 2010; 43: 9–14.
Ridley AJ . Rho GTPases and cell migration. J Cell Sci 2001; 114 (Pt 15): 2713–2722.
Alblas J, Ulfman L, Hordijk P, Koenderman L . Activation of RhoA and ROCK are essential for detachment of migrating leukocytes. Mol Biol Cell 2001; 12: 2137–2145.
Huttenlocher A, Horwitz AR . Integrins in cell migration. Cold Spring Harb Perspect Biol 2011; 3: a005074.
Mitra SK, Schlaepfer DD . Integrin-regulated FAK-Src signaling in normal and cancer cells. Curr Opin Cell Biol 2006; 18: 516–523.
Kalamajski S, Oldberg A . The role of small leucine-rich proteoglycans in collagen fibrillogenesis. Matrix Biol 2010; 29: 248–253.
Oldberg A, Kalamajski S, Salnikov AV, Stuhr L, Morgelin M, Reed RK et al. Collagen-binding proteoglycan fibromodulin can determine stroma matrix structure and fluid balance in experimental carcinoma. Proc Natl Acad Sci USA 2007; 104: 13966–13971.
Kalamajski S, Oldberg A . Fibromodulin binds collagen type I via Glu-353 and Lys-355 in leucine-rich repeat 11. J Biol Chem 2007; 282: 26740–26745.
Lin LC, Hsu SL, Wu CL, Hsueh CM . TGFbeta can stimulate the p(38)/beta-catenin/PPARgamma signaling pathway to promote the EMT, invasion and migration of non-small cell lung cancer (H460 cells). Clin Exp Metastasis 2014; 31: 881–895.
Lu Y, Jiang F, Zheng X, Katakowski M, Buller B, To SS et al. TGF-beta1 promotes motility and invasiveness of glioma cells through activation of ADAM17. Oncol Rep 2011; 25: 1329–1335.
Chopra S, Kumar N, Rangarajan A, Kondaiah P . Context dependent non-canonical WNT signaling mediates activation of fibroblasts by transforming growth factor-beta. Exp Cell Res 2015; 334: 246–259.
Ortuño MJ, Ruiz-Gaspà S, Rodríguez-Carballo E, Susperregui AR, Bartrons R, Rosa JL et al. p38 regulates expression of osteoblast-specific genes by phosphorylation of osterix. J Biol Chem 2010; 285: 31985–31994.
Bevilacqua MA, Iovine B, Zambrano N, D’Ambrosio C, Scaloni A, Russo T et al. Fibromodulin gene transcription is induced by ultraviolet irradiation, and its regulation is impaired in senescent human fibroblasts. J Biol Chem 2005; 280: 31809–31817.
Thillainadesan G, Chitilian JM, Isovic M, Ablack JN, Mymryk JS, Tini M et al. TGF-beta-dependent active demethylation and expression of the p15ink4b tumor suppressor are impaired by the ZNF217/CoREST complex. Mol Cell 2012; 46: 636–649.
Au SL, Wong CC, Lee JM, Wong CM, Ng IO . EZH2-mediated H3K27me3 is involved in epigenetic repression of deleted in liver cancer 1 in human cancers. PLoS One 2013; 8: e68226.
Fernandez AF, Huidobro C, Fraga MF . De novo DNA methyltransferases: oncogenes, tumor suppressors, or both? Trends Genet 2012; 28: 474–479.
Topper JN, DiChiara MR, Brown JD, Williams AJ, Falb D, Collins T et al. CREB binding protein is a required coactivator for Smad-dependent, transforming growth factor beta transcriptional responses in endothelial cells. Proc Natl Acad Sci USA 1998; 95: 9506–9511.
Hildebrand A, Romaris M, Rasmussen LM, Heinegard D, Twardzik DR, Border WA et al. Interaction of the small interstitial proteoglycans biglycan, decorin and fibromodulin with transforming growth factor beta. Biochem J 1994; 302 (Pt 2): 527–534.
Zheng Z, Nguyen C, Zhang X, Khorasani H, Wang JZ, Zara JN et al. Delayed wound closure in fibromodulin-deficient mice is associated with increased TGF-beta3 signaling. J Invest Dermatol 2011; 131: 769–778.
Mayr C, Bund D, Schlee M, Moosmann A, Kofler DM, Hallek M et al. Fibromodulin as a novel tumor-associated antigen (TAA) in chronic lymphocytic leukemia (CLL), which allows expansion of specific CD8+ autologous T lymphocytes. Blood 2005; 105: 1566–1573.
Choudhury A, Derkow K, Daneshmanesh AH, Mikaelsson E, Kiaii S, Kokhaei P et al. Silencing of ROR1 and FMOD with siRNA results in apoptosis of CLL cells. Br J Haematol 2010; 151: 327–335.
Cedar H, Bergman Y, Linking DNA . methylation and histone modification: patterns and paradigms. Nat Rev Genet 2009; 10: 295–304.
Baylin SB . DNA methylation and gene silencing in cancer. Nat Clin Pract Oncol 2005; 2 (Suppl 1): S4–11.
Luo X, Zhang Q, Liu V, Xia Z, Pothoven LK, Lee C . Cutting edge: TGF-β-induced expression of Foxp3 in T cells is mediated through inactivation of ERK. J Immunol 2008; 180: 2757–2761.
Pan X, Chen Z, Huang R, Yao Y, Ma G . Transforming growth factor b1 induces the expression of collagen type I by DNA methylation in cardiac fibroblasts. PLoS One 2013; 8: e60335.
Acknowledgements
The results published here are in whole or part based upon data generated by The Cancer Genome Atlas (TCGA) pilot project established by the NCI and NHGRI. Information about TCGA and the investigators and institutions, which constitute the TCGA research network, can be found at http://cancergenome.nih.gov/. The use of data sets from Institute NC and GSE22866 is acknowledged. IISc NGS facility is acknowledged for methylation and microarray experiments. KS thanks DBT, Government of India for financial support. KS is a JC Bose Fellow of the Department of Science and Technology. BM acknowledges IISc for the fellowship.
Author contributions
KS, VS, AA and ASH coordinated the study; BM and KS conceived and wrote the paper; BM designed, performed and analyzed all the experiments, but with help from MK for experiments shown in Figures 2j, 6f and g, VP for experiments shown in Figures 1 and 7; SDS and KSr performed the experiments shown in Figure 2f. All authors reviewed the results and approved the final version of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Oncogene website
Supplementary information
Rights and permissions
About this article
Cite this article
Mondal, B., Patil, V., Shwetha, S. et al. Integrative functional genomic analysis identifies epigenetically regulated fibromodulin as an essential gene for glioma cell migration. Oncogene 36, 71–83 (2017). https://doi.org/10.1038/onc.2016.176
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/onc.2016.176
This article is cited by
-
Multi-omics sequencing revealed endostar combined with cisplatin treated non small cell lung cancer via anti-angiogenesis
BMC Cancer (2024)
-
A biomechanical view of epigenetic tumor regulation
Journal of Biological Physics (2023)
-
Innovative molecular subtypes of multiple signaling pathways in colon cancer and validation of FMOD as a prognostic-related marker
Journal of Cancer Research and Clinical Oncology (2023)
-
Coupled fibromodulin and SOX2 signaling as a critical regulator of metastatic outgrowth in melanoma
Cellular and Molecular Life Sciences (2022)
-
The role of fibromodulin in cancer pathogenesis: implications for diagnosis and therapy
Cancer Cell International (2019)