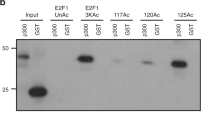
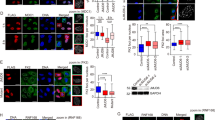
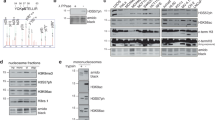
Abstract
The ataxia-telangiectasia mutated (ATM) protein is a key signaling molecule that modulates the DNA damage response. However, the exact mechanism by which ATM regulates DNA damage repair has not yet been elucidated. Here, we report that ATM regulates the DNA damage response by phosphorylating lysine-specific demethylase 2A (KDM2A), a histone demethylase that acts at sites of H3K36 dimethylation. ATM interacts with KDM2A, and their interaction significantly increases in response to DNA double-stranded, but not single-stranded, breaks. ATM specifically phosphorylates KDM2A at threonine 632 (T632) following DNA damage, as demonstrated by a mutagenesis assay and mass spectrometric analysis. Although KDM2A phosphorylation does not alter its own demethylase activity, T632 phosphorylation of KDM2A largely abrogates its chromatin-binding capacity, and H3K36 dimethylation near DNA damage sites is significantly increased. Consequently, enriched H3K36 dimethylation serves as a platform to recruit the MRE11 complex to DNA damage sites by directly interacting with the BRCT2 domain of NBS1, which results in efficient DNA damage repair and enhanced cell survival. Collectively, our study reveals a novel mechanism for ATM in connecting histone modifications with the DNA damage response.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Change history
21 January 2016
This article has been corrected since Advance Online Publication and a corrigendum is also printed in this issue.
References
Kastan MB, Lim DS . The many substrates and functions of ATM. Nat Rev Mol Cell Bio 2000; 1: 179–186.
Shiloh Y . ATM and related protein kinases: safeguarding genome integrity. Nat Rev Cancer 2003; 3: 155–168.
Shiloh Y, Ziv Y . The ATM protein kinase: regulating the cellular response to genotoxic stress, and more. Nat Rev Mol Cell Bio 2013; 14: 197–210.
Burma S, Chen BP, Murphy M, Kurimasa A, Chen DJ . ATM phosphorylates histone H2AX in response to DNA double-strand breaks. J Biol Chem 2001; 276: 42462–42467.
Rogakou EP, Boon C, Redon C, Bonner WM . Megabase chromatin domains involved in DNA double-strand breaks in vivo. J Cell Biol 1999; 146: 905–916.
Guo CY, Wang Y, Brautigan DL, Larner JM . Histone H1 dephosphorylation is mediated through a radiation-induced signal transduction pathway dependent on ATM. J Biol Chem 1999; 274: 18715–18720.
Kim GD, Choi YH, Dimtchev A, Jeong SJ, Dritschilo A, Jung M . Sensing of ionizing radiation-induced DNA damage by ATM through interaction with histone deacetylase. J Biol Chem 1999; 274: 31127–31130.
Pei H, Zhang L, Luo K, Qin Y, Chesi M, Fei F et al. MMSET regulates histone H4K20 methylation and 53BP1 accumulation at DNA damage sites. Nature 2011; 470: 124–128.
Li F, Mao G, Tong D, Huang J, Gu L, Yang W et al. The histone mark H3K36me3 regulates human DNA mismatch repair through its interaction with MutSalpha. Cell 2013; 153: 590–600.
Pfister SX, Ahrabi S, Zalmas LP, Sarkar S, Aymard F, Bachrati CZ et al. SETD2-dependent histone H3K36 trimethylation is required for homologous recombination repair and genome stability. Cell Rep 2014; 7: 2006–2018.
Jha DK, Strahl BD . An RNA polymerase II-coupled function for histone H3K36 methylation in checkpoint activation and DSB repair. Nat Commun 2014; 5: 3965.
Aymard F, Bugler B, Schmidt CK, Guillou E, Caron P, Briois S et al. Transcriptionally active chromatin recruits homologous recombination at DNA double-strand breaks. Nat Struct Mol Biol 2014; 21: 366–374.
Fnu S, Williamson EA, De Haro LP, Brenneman M, Wray J, Shaheen M et al. Methylation of histone H3 lysine 36 enhances DNA repair by nonhomologous end-joining. Proc Natl Acad Sci USA 2011; 108: 540–545.
Wagner EJ, Carpenter PB . Understanding the language of Lys36 methylation at histone H3. Nat Rev Mol Cell Bio 2012; 13: 115–126.
Tsukada Y, Fang J, Erdjument-Bromage H, Warren ME, Borchers CH, Tempst P et al. Histone demethylation by a family of JmjC domain-containing proteins. Nature 2006; 439: 811–816.
Cheng Z, Cheung P, Kuo AJ, Yukl ET, Wilmot CM, Gozani O et al. A molecular threading mechanism underlies Jumonji lysine demethylase KDM2A regulation of methylated H3K36. Genes Dev 2014; 28: 1758–1771.
Tanaka Y, Okamoto K, Teye K, Umata T, Yamagiwa N, Suto Y et al. JmjC enzyme KDM2A is a regulator of rRNA transcription in response to starvation. EMBO J 2010; 29: 1510–1522.
Frescas D, Guardavaccaro D, Kuchay SM, Kato H, Poleshko A, Basrur V et al. KDM2A represses transcription of centromeric satellite repeats and maintains the heterochromatic state. Cell Cycle 2008; 7: 3539–3547.
Wagner KW, Alam H, Dhar SS, Giri U, Li N, Wei Y et al. KDM2A promotes lung tumorigenesis by epigenetically enhancing ERK1/2 signaling. J Clin Invest 2013; 123: 5231–5246.
Lu T, Jackson MW, Singhi AD, Kandel ES, Yang M, Zhang Y et al. Validation-based insertional mutagenesis identifies lysine demethylase FBXL11 as a negative regulator of NF-κB. Proc Natl Acad Sci USA 2009; 106: 16339–16344.
Lu T, Jackson MW, Wang B, Yang M, Chance MR, Miyagi M et al. Regulation of NF-κB by NSD1/FBXL11-dependent reversible lysine methylation of p65. Proc Natl Acad Sci USA 2009; 107: 46–51.
Tewey KM, Rowe TC, Yang L, Halligan BD, Liu LF . Adriamycin-induced DNA damage mediated by mammalian DNA topoisomerase II. Science 1984; 226: 466–468.
Wozniak AJ, Ross WE . DNA damage as a basis for 4'-demethylepipodophyllotoxin-9-(4,6-O-ethylidene-beta-D-glucopyranoside) (etoposide) cytotoxicity. Cancer Res 1983; 43: 120–124.
Zhu W-G, Hileman T, Ke Y, Wang P, Lu S, Duan W et al. 5-Aza-2'-deoxycytidine Activates the p53/p21Waf1/Cip1 Pathway to Inhibit Cell Proliferation. J Biol Chem 2004; 279: 15161–15166.
Cadet J, Sage E, Douki T . Ultraviolet radiation-mediated damage to cellular DNA. Mutat Res 2005; 571: 3–17.
Kim ST, Lim DS, Canman CE, Kastan MB . Substrate specificities and identification of putative substrates of ATM kinase family members. J Biol Chem 1999; 274: 37538–37543.
Matsuoka S, Ballif BA, Smogorzewska A, McDonald ER, Hurov KE, Luo J et al. ATM and ATR Substrate Analysis Reveals Extensive Protein Networks Responsive to DNA Damage. Science 2007; 316: 1160–1166.
Uziel T, Lerenthal Y, Moyal L, Andegeko Y, Mittelman L, Shiloh Y . Requirement of the MRN complex for ATM activation by DNA damage. EMBO J 2003; 22: 5612–5621.
Bakkenist CJ, Kastan MB . DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature 2003; 421: 499–506.
So S, Davis AJ, Chen DJ . Autophosphorylation at serine 1981 stabilizes ATM at DNA damage sites. J Cell Biol 2009; 187: 977–990.
Hickson I, Zhao Y, Richardson CJ, Green SJ, Martin NM, Orr AI et al. Identification and characterization of a novel and specific inhibitor of the ataxia-telangiectasia mutated kinase ATM. Cancer Res 2004; 64: 9152–9159.
Kozlov SV, Graham ME, Peng C, Chen P, Robinson PJ, Lavin MF . Involvement of novel autophosphorylation sites in ATM activation. EMBO J 2006; 25: 3504–3514.
Musselman CA, Kutateladze TG . Handpicking epigenetic marks with PHD fingers. Nucleic Acids Res 2011; 39: 9061–9071.
Lisby M, Rothstein R . DNA damage checkpoint and repair centers. Curr Opin Cell Biol 2004; 16: 328–334.
Stracker TH, Petrini JHJ . The MRE11 complex: starting from the ends. Nat Rev Mol Cell Bio 2011; 12: 90–103.
Lloyd J, Chapman JR, Clapperton JA, Haire LF, Hartsuiker E, Li J et al. A supramodular FHA/BRCT-repeat architecture mediates Nbs1 adaptor function in response to DNA damage. Cell 2009; 139: 100–111.
Xu C, Wu L, Cui G, Botuyan MV, Chen J, Mer G . Structure of a second BRCT domain identified in the nijmegen breakage syndrome protein Nbs1 and its function in an MDC1-dependent localization of Nbs1 to DNA damage sites. J Mol Biol 2008; 381: 361–372.
Oberdoerffer P, Michan S, McVay M, Mostoslavsky R, Vann J, Park SK et al. SIRT1 redistribution on chromatin promotes genomic stability but alters gene expression during aging. Cell 2008; 135: 907–918.
Xie A, Kwok A, Scully R . Role of mammalian Mre11 in classical and alternative nonhomologous end joining. Nat Struct Mol Biol 2009; 16: 814–818.
Rass E, Grabarz A, Plo I, Gautier J, Bertrand P, Lopez BS . Role of Mre11 in chromosomal nonhomologous end joining in mammalian cells. Nat Struct Mol Biol 2009; 16: 819–824.
D'Amours D, Jackson SP . The Mre11 complex: at the crossroads of DNA repair and checkpoint signalling. Nat Rev Mol Cell Bio 2002; 3: 317–327.
Huyen Y, Zgheib O, Ditullio RA Jr, Gorgoulis VG, Zacharatos P, Petty TJ et al. Methylated lysine 79 of histone H3 targets 53BP1 to DNA double-strand breaks. Nature 2004; 432: 406–411.
Sanders SL, Portoso M, Mata J, Bähler J, Allshire RC, Kouzarides T . Methylation of histone H4 lysine 20 controls recruitment of Crb2 to sites of DNA damage. Cell 2004; 119: 603–614.
Pena PV, Davrazou F, Shi X, Walter KL, Verkhusha VV, Gozani O et al. Molecular mechanism of histone H3K4me3 recognition by plant homeodomain of ING2. Nature 2006; 442: 100–103.
Li H, Ilin S, Wang W, Duncan EM, Wysocka J, Allis CD et al. Molecular basis for site-specific read-out of histone H3K4me3 by the BPTF PHD finger of NURF. Nature 2006; 442: 91–95.
Iwase S, Lan F, Bayliss P, de la Torre-Ubieta L, Huarte M, Qi HH et al. The X-linked mental retardation gene SMCX/JARID1C defines a family of histone H3 lysine 4 demethylases. Cell 2007; 128: 1077–1088.
Lan F, Collins RE, De Cegli R, Alpatov R, Horton JR, Shi X et al. Recognition of unmethylated histone H3 lysine 4 links BHC80 to LSD1-mediated gene repression. Nature 2007; 448: 718–722.
Blackledge NP, Zhou JC, Tolstorukov MY, Farcas AM, Park PJ, Klose RJ . CpG islands recruit a histone H3 lysine 36 demethylase. Mol Cell 2010; 38: 179–190.
Zhou JC, Blackledge NP, Farcas AM, Klose RJ . Recognition of CpG island chromatin by KDM2A requires direct and specific interaction with linker DNA. Mol Cell Biol 2011; 32: 479–489.
Wang D, Zhou J, Liu X, Lu D, Shen C, Du Y et al. Methylation of SUV39H1 by SET7/9 results in heterochromatin relaxation and genome instability. Proc Natl Acad Sci USA 2013; 110: 5516–5521.
Khoury-Haddad H, Guttmann-Raviv N, Ipenberg I, Huggins D, Jeyasekharan AD, Ayoub N . PARP1-dependent recruitment of KDM4D histone demethylase to DNA damage sites promotes double-strand break repair. Proc Natl Acad Sci USA 2014; 111: E728–E737.
Young LC, McDonald DW, Hendzel MJ . Kdm4b histone demethylase is a DNA damage response protein and confers a survival advantage following gamma-irradiation. J Biol Chem 2013; 288: 21376–21388.
Mallette FA, Mattiroli F, Cui G, Young LC, Hendzel MJ, Mer G et al. RNF8- and RNF168-dependent degradation of KDM4A/JMJD2A triggers 53BP1 recruitment to DNA damage sites. EMBO J 2012; 31: 1865–1878.
Butler LR, Densham RM, Jia J, Garvin AJ, Stone HR, Shah V et al. The proteasomal de-ubiquitinating enzyme POH1 promotes the double-strand DNA break response. EMBO J 2012; 31: 3918–3934.
Liang CY, Hsu PH, Chou DF, Pan CY, Wang LC, Huang WC et al. The histone H3K36 demethylase Rph1/KDM4 regulates the expression of the photoreactivation gene PHR1. Nucleic Acids Res 2011; 39: 4151–4165.
Liang CY, Wang LC, Lo WS . Dissociation of the H3K36 demethylase Rph1 from chromatin mediates derepression of environmental stress-response genes under genotoxic stress in Saccharomyces cerevisiae. Mol Biol Cell 2013; 24: 3251–3262.
Lee J-H, Paull TT . ATM activation by DNA double-strand breaks through the Mre11-Rad50-Nbs1 complex. Science 2005; 308: 551–554.
Haince JF, McDonald D, Rodrigue A, Dery U, Masson JY, Hendzel MJ et al. PARP1-dependent kinetics of recruitment of MRE11 and NBS1 proteins to multiple DNA damage sites. J Biol Chem 2007; 283: 1197–1208.
Wu L, Luo K, Lou Z, Chen J . MDC1 regulates intra-S-phase checkpoint by targeting NBS1 to DNA double-strand breaks. Proc Natl Acad Sci USA 2008; 105: 11200–11205.
Celeste A, Petersen S, Romanienko PJ, Fernandez-Capetillo O, Chen HT, Sedelnikova OA et al. Genomic Instability in Mice Lacking Histone H2AX. Science 2002; 296: 922–927.
Kobayashi J, Tauchi H, Sakamoto S, Nakamura A, Morishima K, Matsuura S et al. NBS1 localizes to gamma-H2AX foci through interaction with the FHA/BRCT domain. Curr Biol 2002; 12: 1846–1851.
Hari FJ, Spycher C, Jungmichel S, Pavic L, Stucki M . A divalent FHA/BRCT-binding mechanism couples the MRE11-RAD50-NBS1 complex to damaged chromatin. EMBO Rep 2010; 11: 387–392.
Wang Q, Goldstein M, Alexander P, Wakeman TP, Sun T, Feng J et al. Rad17 recruits the MRE11-RAD50-NBS1 complex to regulate the cellular response to DNA double-strand breaks. EMBO J 2014; 33: 862–877.
Huang Y, Liu Y, Yu L, Chen J, Hou J, Cui L et al. Histone demethylase KDM2A promotes tumor cell growth and migration in gastric cancer. Tumour Biol 2014; 36: 271–278.
Wang H, Zhou W, Zheng Z, Zhang P, Tu B, He Q et al. The HDAC inhibitor depsipeptide transactivates the p53/p21 pathway by inducing DNA damage. DNA Repair (Amst) 2012; 11: 146–156.
Acknowledgements
We thank Dr David J Chen at University of Texas Southwestern Medical Center for kindly providing YFP-ATM plasmid. We also thank Drs Jun Huang at Zhejiang University and Jiadong Wang at Peking University Health Science Center for kindly providing NBS1 deleted mutants. This work was supported by National Key Basic Research Program of China grant 2011CB504200, 2013CB911000; National Natural Science Foundation of China grants 31070691, 81321003, 91319302; Minister of Education of China ‘111 Project’.
Author information
Authors and Affiliations
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Oncogene website
Supplementary information
Rights and permissions
About this article
Cite this article
Cao, LL., Wei, F., Du, Y. et al. ATM-mediated KDM2A phosphorylation is required for the DNA damage repair. Oncogene 35, 301–313 (2016). https://doi.org/10.1038/onc.2015.81
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/onc.2015.81
This article is cited by
-
A novel STAT3/ NFκB p50 axis regulates stromal-KDM2A to promote M2 macrophage-mediated chemoresistance in breast cancer
Cancer Cell International (2023)
-
BPA induces placental trophoblast proliferation inhibition and fetal growth restriction by inhibiting the expression of SRB1
Environmental Science and Pollution Research (2023)
-
Biological function and regulation of histone 4 lysine 20 methylation in DNA damage response
Genome Instability & Disease (2022)
-
Histone methyltransferase GLP epigenetically activates GPCPD1 to sustain cancer cell metastasis and invasion
Genome Instability & Disease (2022)
-
1H NMR-based assay for lysine demethylase LSD1 and its application to inhibitor screening
Genome Instability & Disease (2021)