Abstract
Whole-genome and transcriptome sequencing were used to discover novel gene fusions in a case of colon cancer. A tumor-specific LACTB2-NCOA2 fusion originating from intra-chromosomal rearrangement of chromosome 8 was identified at both DNA and RNA levels. Unlike conventional oncogenic chimeric proteins, the fusion product lacks functional domain from respective genes, indicative of an amorphic rearrangement. This chimeric LACTB2-NCOA2 transcript was detected in 6 out of 99 (6.1%) colorectal cancer (CRC) cases, where NCOA2 was significantly downregulated. Enforced expression of wild-type NCOA2 but not the LACTB2-NCOA2 fusion protein impaired the pro-tumorigenic phenotypes of CRC cells, whereas knockdown of endogenous NCOA2 in normal colonocytes had opposite effects. Mechanistically, NCOA2 inhibited Wnt/β-catenin signaling through simultaneously upregulating inhibitors and downregulating stimulators of Wnt/β-catenin pathway. Collectively, our data supports that NCOA2 is a novel negative growth regulatory gene repressing the Wnt/β-catenin pathway in CRC, where recurrent fusion with LACTB2 contributes to its disruption.
Similar content being viewed by others
Introduction
Colorectal cancer (CRC) is the third most commonly diagnosed cancer and the fourth leading cause of cancer death globally. Each year over one million people are diagnosed with CRC worldwide with a mortality rate of ~30%.1 The molecular pathogenesis of sporadic CRC is complex and heterogeneous, involving somatic mutations of a constellation of oncogenes (for example, KRAS, PIK3CA and BRAF) and tumor suppressor genes (for example, APC, TP53 and FBXW7).2 These acquired genetic defects cause hyperactivation or inactivation of different intracellular signals, resulting in deregulated phenotypes, such as uncontrolled cell proliferation, resistance to cell death, and cellular invasiveness and metastasis, thereby contributing to colorectal tumorigenesis.3 With the recent advent of next-generation sequencing, identification of genetic aberrations in CRC has been advanced to genome-wide level. Apart from abovementioned CRC hallmark genes, a large number of novel candidates (for example, ARID1A, SOX9 and FAM123B) mutated at lower frequencies in CRC have been unveiled by high-throughput sequencing with anticipated functional impacts on diverse cellular processes, such as chromatin remodeling and stemness.4 Through exome and targeted capture sequencing, our group recently reported additional recurrently mutated genes (that is, CDH10, FAT4 and DOCK2) that are known regulators of canonical and non-canonical Wnt signaling, in the Asian CRC cohort.5
Aside from microscale alterations in DNA sequence (that is, point mutations and small insertions/deletions), next-generation sequencing possesses the power to identify gross genetic defects, including abnormal gene fusions originated from chromosomal rearrangements.6, 7 Such fusion events could promote tumorigenesis by mediating proto-oncogene overexpression via promoter exchange (for example, Ig-MYC in multiple myeloma) or disruption of microRNA-binding site (for example, FGFR3-TACC3 in bladder cancer and glioblastoma).8, 9, 10 Gene fusions could also produce constitutively active chimeric proteins (for example, BCR-ABL1 in chronic myeloid leukemia and acute lymphoblastic leukemia) and inactivate tumor suppressor genes (for example, fusion of TTC28 with multiple loci in CRC).4, 7, 11 Although most gene fusions were originally described in hematological malignancies, the application of next-generation sequencing has led to the confirmation of their pervasive existence in solid tumors.7 Thus far, several fusion transcripts, namely C2orf44-ALK, VTI1A-TCF7L2, NAV2-TCF7L1, EIF3E-RSPO2 and PTPRK-RSPO3, of various prevalence (2.5–7.4%) have been documented in CRC.4, 12, 13, 14 In particular, the functional relevance of R-spondin fusion proteins (that is, EIF3E-RSPO2 and PTPRK-RSPO3) to the activation of oncogenic Wnt/β-catenin signaling has been demonstrated.12
In the present study, we adopted an integrative approach for the discovery of novel gene fusions in CRC. We first performed whole-genome and transcriptome sequencing in a case of CRC to identify fusion events that could be detected at both DNA and RNA levels. Large-cohort validation followed by in vitro and in vivo functional assays were then used to assess the prevalence and biological significance of identified fusion events. With these efforts, we successfully identified a recurrent fusion that disrupts the novel negative growth regulatory gene NCOA2 in CRC.
Results
Whole-genome and transcriptome sequencing identified novel gene fusions in CRC
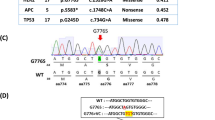
To identify fusion events in CRC on a genome-wide scale, we performed transcriptome and whole-genome sequencing of a microsatellite-stable CRC sample staged at T3N1M1, with adjacent normal tissue and peripheral blood as normal control (Supplementary Table 1). Examination of the discordant paired-end reads at RNA level revealed that 13 chimeric transcripts contained in-frame exons from separate autosomal genes (Supplementary Table 2). In parallel, a total of 193 somatic structural variations (SVs) were identified at DNA level, of which 130 (67.4) and 63 (32.6%) were primarily intra-chromosomal and inter-chromosomal, respectively, with 128 displayed interruption of gene-coding regions (Figure 1a; Supplementary Table 3). To screen for novel SV-based fusion of potential significance, we selected the chimeric genes with consistent breakpoints at both RNA and DNA levels, and removed known fusions, which are present in public databases such as ChimeraDB. After filtering, we discovered three tumor-specific fusions derived from chromosomal translocations. The first two fusions were TM9SF2-13q31 (two transcript variants) and TRAM1-8q22 (three transcript variants) (Supplementary Figure 1), both of which have similar structures that gene-coding regions were disrupted by non-coding intergenic fragments, resulting in gene truncation. On the other hand, the third one was a fusion involving the 1st exon of LACTB2 and the 22nd exon of NCOA2 (Figure 1b). Of note, the identified chimeric DNA structure of this fusion comprised an additional non-coding intron fragment of VPS13B between the expected junctions of LACTB2 and NCOA2, indicating that the hybrid LACTB2-NCOA2 transcript was originated from structurally rearranged chromosomes.
Recurrent LACTB2-NCOA2 fusion in colorectal cancer (CRC). (a) Structural variations (SVs) in the tumor tissue of a CRC case were illustrated as a Circos plot with red and blue lines indicating inter-chromosomal and intra-chromosomal SVs, respectively. (b) The DNA and RNA structures of the LACTB2-NCOA2 fusion were illustrated. RT–PCR (M: size marker; N: normal cDNA; T: CRC cDNA) and Sanger sequencing confirmed the tumor-specific occurrence of LACTB2-NCOA2 fusion. (c) The LACTB2-NCOA2 fusion transcript with the same breakpoints was present in 6 out of 99 (6.1%) CRC cDNA samples as confirmed by RT–PCR and Sanger sequencing (chromatogram).
LACTB2-NCOA2 fusion was a recurrent, tumor-specific event in CRC
Next, we assessed the molecular prevalence of these three identified fusions in a large CRC cohort by PCR. Despite strenuous efforts to target different breakpoint sequences, we failed to detect the recurrence of TM9SF2-13q31 fusion in CRC complementary DNA (cDNA; data not shown), indicating that this fusion is a rare event with limited impact. Conversely, the TRAM1-8q22 fusion was present in both tumor and normal cDNA (data not shown), suggesting that this chimera was not tumor-specific. Intriguingly, the LACTB2-NCOA2 fusion was present in 6 out of 99 (6.1%) CRC cDNA samples were with the same breakpoints (Figure 1c), whereas no similar hybrid was detected in the normal counterparts. Further sequence analysis showed that, in stark contrast to classical oncogenic gene fusions which often retain the key structures of individual genes to confer growth and/or survival advantages to tumor,12, 13, 14 the LACTB2-NCOA2 fusion only encoded truncated non-domain fragments, suggestive of an amorphic rearrangement in which LACTB2 and/or NCOA2 might be a tumor suppressor silenced by chromosomal translocation in CRC.
NCOA2 was downregulated in CRC
A common feature of functional negative growth regulatory genes is their downregulation in cancer. We therefore measured the expression of LACTB2 and NCOA2 in paired CRC tissues. Quantitative reverse transcription–PCR (qRT–PCR) revealed that NCOA2, but not LACTB2, was evidently downregulated in CRC tissues compared with the cancer-adjacent controls (Figure 2a). Further immunohistochemical analysis of paired CRC tissues confirmed significantly weaker nuclear expression of NCOA2 in tumor cells than in normal colonocytes (Figure 2b), concordant with the reported function of NCOA2 as a transcriptional regulator.15, 16 The specificity of NCOA2 antibody was confirmed by performing immunocytochemistry using SW1116 cells transfected with empty (pcDNA3.1) or NCOA2-encoding plasmid. Enforced expression of NCOA2 markedly increased the staining intensity with the antibody (Figure 2c). Moreover, qRT–PCR demonstrated that NCOA2 expression was markedly lowered in CRC cell lines compared with normal colorectal tissues (Figure 2d).
Downregulation of NCOA2 in CRC. (a) The mRNA expression levels of NCOA2, but not LACTB2, were significantly lower in CRC tissues compared with corresponding tumor-adjacent controls as measured by qRT–PCR. (b) Immunohistochemical staining revealed significantly lower NCOA2 protein expression in CRC tissues as compared with corresponding tumor-adjacent tissues. (c) Immunoreactivity of NCOA2 was significantly higher in SW1116 cells overexpressing NCOA2. (d) NCOA2 mRNA levels were much lower in CRC cell lines compared with normal colonic tissues. Results from qRT–PCR and conventional RT–PCR are shown in the chart (top) and as a gel picture (bottom), respectively. Representative micrographs and quantification results of three independent experiments are shown.
NCOA2 but not LACTB2-NCOA2 fusion exerted tumor-suppressive functions
To directly compare the functional impact of wild-type NCOA2 with that of the fusion protein in CRC, we performed multiple in vitro assays on HCT116 (p53 wildtype) and SW1116 (p53 mutant) colon cancer cells stably transfected with empty vector (control), vector encoding full-length NCOA2 or vector encoding the LACTB2-NCOA2 fusion protein (Figure 3a). Results showed that enforced expression of wild-type NCOA2 strongly inhibited the proliferation (Figure 3b) and colony formation (Figure 3c) of both HCT116 and SW1116 cells, whereas the LACTB2-NCOA2 fusion exerted no significant effect under the same conditions. Annexin V staining, cell migration and invasion assays further revealed that NCOA2 but not the LACTB2-NCOA2 fusion protein increased early apoptosis (Figure 3d), impeded migration and suppressed invasiveness (Figure 3e) in both CRC cell lines. Consistent with in vitro findings, overexpression of NCOA2 but not the LACTB2-NCOA2 fusion protein significantly inhibited the growth of HCT116 xenografts in nude mice (Figure 3f). The overexpression of NCOA2 in xenografts established from NCOA2-stably transfected HCT116 cells was confirmed by immunohistochemistry (Supplementary Figure 2).
Tumor-suppressing functions of NCOA2 and their abrogation after fusion with LACTB2 (a) The mRNA expression of full-length NCOA2 and the LACTB2-NCOA2 fusion (LN-fusion) in stably transfected HCT116 and SW1116 cells was confirmed by RT–PCR. The protein expression of full-length NCOA2 after stable transfection was also confirmed by Western blots (WB). (b–e) Enforced expression of full-length NCOA2, but not the LN-fusion, significantly (b) lowered proliferation as measured by xCELLigence System, (c) impaired colony formation, (d) induced early apoptosis and (e) inhibited migration (left) and invasion (right) of HCT116 and SW1116 cells. (f) The growth of HCT116 xenografts stably expressing NCOA2 was significantly slower than those stably transfected with empty vector or LN-fusion in nude mice. Data are represented as mean±s.d. of three separate experiments in triplicates. *P<0.05; **P<0.01; ***P<0.005 significantly different between indicated groups.
Knockdown of NCOA2 promoted oncogenic phenotypes in normal colonocytes
To confirm the tumor-suppressing action of NCOA2, we knocked down the expression of this gene using two different small-interfering RNA (siRNA) sequences in normal colonocytes NCM460 (Figure 4a). Functional analyses showed that depletion of endogenous NCOA2 markedly increased cell proliferation (Figure 4b) and reduced early apoptosis (Figure 4c) of NCM460, which were in line with the results from gain-of-function studies of NCOA2.
Pro-tumorigenic effects of NCOA2 knockdown in normal colonocytes NCM460. (a) The downregulation of mRNA and protein expression of endogenous NCOA2 on transfection with siRNA was confirmed by RT–PCR and Western blots (WB), respectively. (b) Proliferation of NCOA2-knocked down NCM460 cells was accelerated when compared with control siRNA-transfected cells. (c) Basal apoptosis of NCM460 cells was significantly reduced on transfection with NCOA2 siRNA. Assays were conducted 48 h post transfection and data are represented as mean±s.d. of three separate experiments in triplicates. *P<0.05; **P<0.01 significantly different between indicated groups.
Wild-type NCOA2 but not LACTB2-NCOA2 fusion repressed Wnt signaling
Previous studies have shown that NCOA2 could act as a co-activator of nuclear receptors, such as vitamin D receptor15 and retinoic acid receptor,16 which are known suppressors of Wnt/β-catenin signaling.17, 18, 19 Therefore, to gain mechanistic insights into the oncogenic role of disruptive LACTB2-NCOA2 fusion, we performed Wnt signaling-dependent luciferase reporter assays in HCT116 cells. Compared with the empty vector control, wild-type NCOA2 significantly reduced the protein levels of active β-catenin (Figure 5a) and Wnt-dependent luciferase activity (Figure 5b), whereas the LACTB2-NCOA2 fusion exerted negligible effects. Enforced expression of wild-type NCOA2 but not the fusion protein also deregulated the expression of a repertoire of Wnt signaling regulators (Figure 5c). In particular, restored expression of NCOA2 simultaneously upregulated inhibitors (for example, APC and Frzb) and downregulated stimulators (for example, Frizzled, Dishevelled and casein kinase 1) of the Wnt signaling pathway. The repression of Wnt/β-catenin signaling by NCOA2 was also confirmed by the downregulation of FOSL1 and Jun, both of which are transcriptional targets of β-catenin/TCF complex (Figure 5d; Supplementary Table 4).
Repression of Wnt/β-catenin by wild-type NCOA2 but not the LACTB2-NOCA2 fusion (LN-fusion) protein in CRC cells. (a and b) Western blots and luciferase reporter activity show (a) the reduction of active β-catenin and (b) the inhibition of β-catenin transcriptional activity on enforced expression of NCOA2, but not the LN-fusion protein, in stably transfected HCT116 cells. (c) Stable overexpression of NCOA2, but not the LN-fusion protein, caused dramatic alterations in the expression of Wnt signaling components in HCT116 cells as measured by pathway-specific RT–PCR array. (d) The concurrent upregulation of Wnt inhibitors (for example, APC and Frzb) and downregulation of Wnt stimulators (for example, Frizzled, CK1 and Dvl) as well as the repression of β-catenin transcriptional targets (that is, FOSL1 and Jun) on stable expression of NOCA2 in HCT116 cells were illustrated in the schematic diagram. Gel pictures are representative and data are represented as mean±s.d. of three independent experiments. *P<0.05 significantly different between indicated groups.
Discussion
Using state-of-the-art sequencing technologies, we interrogated the genome and transcriptome of a case of CRC in which three SVs that give rise to concordant fusion transcripts were identified. Further validation in our CRC cohort confirmed that one of these three fusions, namely LACTB2-NCOA2, was a tumor-specific, recurrent event. The fusion protein lacks major functional domains of respective genes, indicative of a loss-of-function rearrangement. Subsequent analyses revealed that NCOA2 but not LACTB2 was downregulated in CRC in which wild-type NCOA2 but not the LACTB2-NCOA2 fusion protein exerted tumor-suppressing functions, including induction of apoptosis, inhibition of cell motility and invasiveness, and reduction of in vivo tumorigenicity. Mechanistic studies showed that NCOA2 but not the fusion protein inhibited Wnt signaling through concordant deregulation of Wnt regulators. Collectively, our data substantiated the notion that the destructive fusion with LACTB2 disrupted the tumor-suppressing functions of NCOA2 thereby promoting colorectal tumorigenesis. However, it must be pointed out that the function of NCOA2 in tumorigenesis is context dependent. For instance, NCOA2 has been shown to be pro-tumorigenic in brain and prostate cancers.20, 21 Such functional divergence may be attributed to the different molecular pathways regulated by NCOA2 in individual cancers. The ability of NCOA2 to differentially modulate the expression of inhibitors and stimulators of Wnt signaling may render its role as a tumor suppressor in CRC.
In this case of whole-genome-sequenced CRC, the number of intra-chromosomal translocation far exceeds that of inter-chromosomal translocation. The former most often results from a segment breaking off the involved chromosome and being rejoint at a different location. In contrast, inter-chromosomal translocation is frequently mediated by non-allelic homologous recombination.22 However, we observed that most intra-chromosomal translocations are clustered on chromosome 8 (Figure 1a). Such massive genomic rearrangement, also known as chromothrypsis, usually occurs after a single catastrophic event leading to chromosome fragmentation followed by aberrant repair. It has been reported that as many as 3% of all cancers exhibit chromothrypsis.23 Nevertheless, the underlying molecular mechanism and its relation to LACTB2-NCOA2 fusion (an intra-chromosomal translocation in chromosome 8) remain elusive.
Previous studies have established the role of NCOA2 as a crucial co-activator of diverse nuclear receptors, which regulate a wide range of physiological responses.16, 24, 25, 26 Among the nine NCOA2-associated nuclear receptors, estrogen receptor, retinoic acid receptor/retinoid X receptor, thyroid receptor and vitamin D receptor are of particular relevance to CRC as activation of these receptors has been shown to suppress CRC growth and induce cell differentiation and apoptosis.27 Moreover, these particular nuclear receptors are potent suppressors of Wnt/β-catenin signaling.18 Our finding that NCOA2 attenuates CRC growth, particularly its robust inhibitory effects on Wnt signaling, is strongly coherent with the reported tumor-suppressing functions of its associated nuclear receptors in CRC.
In human, chimeric transcripts are known to be generated via different biogenic pathways, including trans-splicing of pre-mRNAs,28 RNA transcription runoff,29 RNA processing errors28 and gene fusion due to intra- or inter-chromosomal translocations.14, 30, 31 In our study, the presence of an additional non-coding intron fragment of VPS13B (also on chromosome 8) between LACTB2 and NCOA2 at DNA level strongly indicated that the LACTB2-NCOA2 fusion was generated through complex intra-chromosomal rearrangement. Moreover, the absence of its detection in normal tissues confirmed that the LACTB2-NCOA2 fusion is a tumor-specific somatic event. Given that LACTB2-NCOA2 fusion only occurs in a small subset (~6%) of CRC cases, whereas NCOA2 exhibited pervasive downregulation, mechanisms other than gene fusion should have been involved in NCOA2 inactivation. In this regard, several microRNAs upregulated in CRC (for example, miR-181b, miR-200c and miR-1290)32, 33 were predicted to target NCOA2 by multiple computer algorithms, namely miRanda, miRDB, miRWalk, PICTAR5 and Targetscan. NCOA2 also harbors CpG islands in its promoter,34 rendering the possibility of NCOA2 regulation by microRNAs and/or DNA hypermethylation.
According to a recent systematic analysis, hundreds of chimeric RNAs exist in different human tissues and certain chimeric products are highly tissue specific with functional implications.35 While the physiological significance of most identified chimeric transcripts remains elusive, increasing evidence supports a strong correlation between certain chimeric proteins and human disorders, particularly cancers. With the advent of high-throughout sequencing technology, increasing number of fusion transcripts is being described in malignancies, with the majority of fusion events resulting in the generation of oncogenic chimeric proteins.7 Aside from activation of proto-oncogene, gene fusion could mediate their biological functions through gene disruption (for example, PAFAH1B3-CLK2 in mental retardation and TTC28 in CRC).4, 36 However, the disruption of tumor-suppressor gene through recurrent in-frame fusion of two genes in cancer has not yet been described. Herein, we report the first of its kind by showing that wild-type NCOA2 but not the in-frame fusion product possessed tumor-suppressing functions. Importantly, although fusions of NCOA2 with other genes have been reported in human cancers (for example, PAX3-NCOA2 in rhabdomyosarcoma,37 HEY1-NCOA2 in mesenchymal chondrosarcoma38 and GTF2I-NCOA2 in soft tissue angiofibroma),39 these NCOA2 chimeras retain the functional domains of NCOA2, whose biological function is presumed to be augmented by its fusion partner to induce carcinogenic transformation.40, 41, 42
In conclusion, we demonstrated an unreported function of recurrent in-frame gene fusion in promoting colorectal carcinogenesis through inactivation of the negative growth regulatory gene NCOA2 in CRC. Given the enormous amount of chimeric sequences in cancer genomes, it is anticipated that a large number of loss-of-function fusion events remain unexplored. Future identification and characterization of such fusions using revised screening strategies may uncover a new repertoire of growth regulators with clinical and functional significance.
Materials and methods
Human samples and DNA/RNA extraction
A histologically confirmed, moderately differentiated adenocarcinoma isolated from the distal transverse colon of a 65-year-old Chinese male admitted to the Prince of Wales Hospital in 2010 was subject to genomic analysis. The collected tumor was staged as T3N1M1 according to the tumor-node-metastasis (TNM) system by a pathologist and was classified as microsatellite stable based on the absence of mutation in all five Bethesda markers (BAT25, BAT26, D2S123, D5S346 and D17S250). Written consent had been obtained prior to collection of tumor, adjacent normal tissue and peripheral blood samples. Clinical samples for prevalence screening were colon tumors and adjacent normal tissue obtained from surgical resections in the Prince of Wales Hospital in the past decade. Written consents had been obtained from all patients involved. Genomic DNA was extracted from tumor and blood samples using QIAamp DNA Mini Kit (Qiagen, Valencia, CA, USA). Total RNA was isolated from tissue samples and cancer cell lines using Trizol reagent (Invitrogen, Carlsbad, CA, USA) following manufacturer’s instructions. This study had been approved by the Clinical Research Ethics Committee of the Chinese University of Hong Kong.
Cell lines and cell culture
The human colon cancer cell lines CaCO2, DLD-1, HCT116, LoVo, LS180, SW480, SW620 and SW1116 were purchased from the American Type Culture Collection (Manassas, VA, USA). The human colon cancer cell line CL-14 was purchased from Creative Bioarray (New York, NY, USA). All cell lines had been authenticated with short-tandem repeat profiling by the vendors. Cells were cultured in RPMI-1640, Dulbecco's Modified Eagle's medium (DMEM) or Eagle's medium (Invitrogen) supplemented with 10% (v/v) fetal bovine serum (Invitrogen).
Transcriptome sequencing and identification of fusion RNAs
Total RNA isolated from the primary colon tumor tissue was subject to transcriptome sequencing. Poly-A-containing mRNA purification, double-stranded cDNA synthesis, end repair, 3’ end adenylation, adapter ligation and enrichment of DNA fragments for RNA-Seq library construction were performed using the reagents provided in the Illumina TruSeq RNA Sample Preparation Kit (Illumina, San Diego, CA, USA). RNA-Seq library sequencing was then performed on an Illumina HiSeq 2000 (Illumina) as per manufacturer’s instructions. The sequencing reads were first filtered to remove low quality reads or reads with adapters. All remaining qualified reads were aligned to human hg19 downloaded from UCSC (Santa Cruz, CA, USA). In total, 11.5 gb qualified sequence reads were obtained, of which ~71% mapped uniquely to the human genome. The fragments per kb of exon per million fragments mapped (FPKM) expression levels for each gene were calculated using the program Cufflinks. Both SOAPfuse (http://soap.genomics.org.cn/soapfuse.html) and deFuse programs (Vancouver, BC, Canada) were used for scanning of fusion RNAs using transcriptome data. The de novo transcriptome assembler SOAPdenovo-Trans was then used for transcript assembly to double check gene fusions matching somatic SVs (http://soap.genomics.org.cn/SOAPdenovo-Trans.html).
Whole-genome sequencing and SV calling
Genomic DNA from primary tumor and peripheral blood was fragmented to an average size of 500 nucleotides. Standard Illumina protocols (Illumina) and Illumina paired-end adapters (Illumina) were then used for library preparation. DNA library sequencing was then performed on an Illumina Solexa sequencing platform (Illumina) as per manufacturer’s instructions. The general clustering diagram was used to call SVs similar to dRanger (Cambridge, MA, USA). First, fragment-length distribution was analyzed to evaluate the library insert size range and filter discordant read pairs, which refer to mapping to different chromosomes or in unexpected positions (>max library insert size+4.5 × s.d.) or unexpected orientations (incorrect order on opposite strands or any order on the same strand) on the same chromosome. Second, clusters of discordant pairs implicated potential rearrangements and determined the rough range of SVs. To filter out somatic SVs, clusters with any supporting discordant pairs in the same region from matched normal were discarded. Moreover, clusters with less than five supporting read pairs or falling to UCSC simple Repeat regions were discarded. To identify the exact SV breakpoints at single nucleotide level, an in-house program SeekSV was used. Similar to CREST, SeekSV used next-generation short reads with partial alignments with the reference genome to call SVs and included four steps as follows: (i) obtain soft-clipped reads from the BWA alignment results; (ii) align the clipped sequences (unmapped parts of the soft-clipped reads) with the human reference genome hg19; (iii) obtain the SV breakpoints according to the break end positions in the alignment results; (iv) obtain somatic SV breakpoints by comparing SVs in tumor with those in blood. SV breakpoints identified by SeekSV with concordant SV clusters predicted by the clustering method above were then selected to fill the SV gaps. Sequences disrupted by SVs were annotated to gene regions according to refGene database downloaded from UCSC.
Validation and screening for gene fusion
Expression of indicated RNA fusions was validated in CRC tumor and adjacent normal samples by RT–PCR. Sample cDNA was synthesized using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA, USA). RT–PCR was performed using AmpliTaq Gold DNA Polymerase (Invitrogen) or MightyAmp DNA Polymerase (Takara, Dalian, China) as per manufacturer’s instructions.
Examination of NCOA2 expression
NCOA2 expression levels were examined in tissue samples and cell lines by qRT–PCR and/or immunohistochemistry. qRT–PCR was performed on LightCycler 480 real-time PCR system (Roche Applied Science, Indianapolis, IN, USA) using LightCycler 480 SYBR Green I Master (Roche Applied Science) following manufacturer’s instructions. Immunohistochemistry was performed on paraffin sections using an anti-NCOA2 antibody (Cat: sc-8996; Santa Cruz Biotechnology, Santa Cruz, CA, USA).
Construction and transfection of expression vector/siRNA
The expression vectors were constructed by inserting the putative open reading frames (ORF) encoded by the LACTB2-NCOA2 fusion RNA and the full-length ORF of human NCOA2 into the pcDNA3.1-V5-His-TOPO expression vector (Invitrogen). The sequences of the constructs were confirmed by Sanger sequencing. Knockdown of NCOA2 was mediated by custom-made siRNA (GenePharma, Shanghai, China). Transfection of vector and siRNA was performed using Lipofectamine 2000 (Invitrogen) following manufacturer’s instructions. Stably transfected cells were established under selection with neomycin (G418) (Invitrogen). Ectopic expression of target genes was confirmed by RT–PCR and/or Western blot. RNA and proteins were harvested at 48 h after transfection.
Western blots
Total protein was extracted and protein concentration was measured by the DC protein assay method of Bradford (Bio-Rad, Hercules, CA, USA). Protein samples (30 μg per lane) were separated on 10% Bis/Tris-polyacrylamide gel by electrophoresis and blotted onto nitrocellulose membranes (GE Healthcare, Piscataway, NJ, USA). Blots were stained with primary antibodies overnight at 4 °C and secondary antibodies for 1 h at room temperature. Proteins were visualized using ECL Plus Western blotting Detection Reagents (GE Healthcare).
Cell proliferation and colony formation assays
Cell proliferation was monitored real-time by an xCELLigence System (Roche Applied Science) with growth index data collected automatically every 30 min until the chamber becomes saturated with cells or measured by CellTiter 96 AQueous One Solution Cell Proliferation Assay (Promega, Madison, WI, USA) according to manufacturer’s instructions. For colony formation, cells were seeded (2 × 103 per well) onto 6-well plate for 10–14 days, resulting colonies (>50 cells) were then counted after being fixed with 70% ethanol and stained with crystal violet solution.
Flow cytometry for apoptosis
Cells were stained with Annexin V (allophycocyanin conjugated) and 7-AAD (BD Biosciences), then analyzed by fluorescence-activated cell sorting using FACScan (BD Biosciences, San Jose, CA, USA). Cell populations were classified as viable (Annexin V negative, 7-AAD negative), early apoptotic (Annexin V positive, 7-AAD negative), late apoptotic (Annexin V positive, 7-AAD positive) or necrotic (Annexin V negative, 7-AAD positive).
In vivo tumorigenicity assays
HCT116 cells (1.5 × 106 cells) transfected with empty vector, pcDNA3.1-LACTB2(e1)-NCOA2(e22) or pcDNA3.1-NCOA2 were injected subcutaneously into dorsal flanks of 4-week-old female Balb/c nude mice (n=5 per group) on day 0. Tumor diameters were measured every 2 days for ~2 weeks. Tumor volume (mm3) was estimated by measuring the longest and shortest diameter of the tumor. All experimental procedures had been approved by the Animal Ethics Committee of the Chinese University of Hong Kong.
Wnt signaling pathway-PCR array
Gene expression profiles of stably transfected HCT116 cells were analyzed by Human Wnt Signaling Pathway RT2 Profiler PCR Array (SABiosciences, Frederick, MD, USA). Each array contains 84 functionally characterized genes related to Wnt-mediated signal transduction (http://www.sabiosciences.com). Gene expression with more than threefold of change was considered to be of biological significance.
Statistical analysis
Results were expressed as mean±s.d. Statistical analysis was performed using the SPSS statistical software package (standard version 13.0; IBM China/Hong Kong Limited, Hong Kong, China). Mann–Whitney U-test or Student’s t-test was used to compare the variables of two groups. Analysis of variance was used for multiple-group comparisons. P-values<0.05 were taken as statistically significant.
References
Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D . Global cancer statistics. CA Cancer J Clin 2011; 61: 69–90.
Wong SH, Sung JJ, Chan FK, To KF, Ng SS, Wang XJ et al. Genome-wide association and sequencing studies on colorectal cancer. Semin Cancer Biol 2013; 23: 502–511.
Wu WK, Wang XJ, Cheng AS, Luo MX, Ng SS, To KF et al. Dysregulation and crosstalk of cellular signaling pathways in colon carcinogenesis. Crit Rev Oncol Hematol 2013; 86: 251–277.
Cancer Genome Atlas Network. Comprehensive molecular characterization of human colon and rectal cancer. Nature 2012; 487: 330–337.
Yu J, Wu WK, Li X, He J, Li XX, Ng SS et al. Novel recurrently mutated genes and a prognostic mutation signature in colorectal cancer. Gut 2014; pii: gutjnl-2013-306620.
Bunting SF, Nussenzweig A . End-joining, translocations and cancer. Nat Rev Cancer 2013; 13: 443–454.
Annala MJ, Parker BC, Zhang W, Nykter M . Fusion genes and their discovery using high throughput sequencing. Cancer Lett 2013; 340: 192–200.
Guo G, Sun X, Chen C, Wu S, Huang P, Li Z et al. Whole-genome and whole-exome sequencing of bladder cancer identifies frequent alterations in genes involved in sister chromatid cohesion and segregation. Nat Genet 2013; 45: 1459–1463.
Parker BC, Annala MJ, Cogdell DE, Granberg KJ, Sun Y, Ji P et al. The tumorigenic FGFR3-TACC3 gene fusion escapes miR-99a regulation in glioblastoma. J Clin Invest 2013; 123: 855–865.
Dib A, Gabrea A, Glebov OK, Bergsagel PL, Kuehl WM . Characterization of MYC translocations in multiple myeloma cell lines. J Natl Cancer Inst Monogr 2008; 2008: 25–31.
Quintas-Cardama A, Cortes J . Molecular biology of bcr-abl1-positive chronic myeloid leukemia. Blood 2009; 113: 1619–1630.
Seshagiri S, Stawiski EW, Durinck S, Modrusan Z, Storm EE, Conboy CB et al. Recurrent R-spondin fusions in colon cancer. Nature 2012; 488: 660–664.
Lipson D, Capelletti M, Yelensky R, Otto G, Parker A, Jarosz M et al. Identification of new ALK and RET gene fusions from colorectal and lung cancer biopsies. Nat Med 2012; 18: 382–384.
Bass AJ, Lawrence MS, Brace LE, Ramos AH, Drier Y, Cibulskis K et al. Genomic sequencing of colorectal adenocarcinomas identifies a recurrent VTI1A-TCF7L2 fusion. Nat Genet 2011; 43: 964–968.
Takeyama K, Masuhiro Y, Fuse H, Endoh H, Murayama A, Kitanaka S et al. Selective interaction of vitamin D receptor with transcriptional coactivators by a vitamin D analog. Mol Cell Biol 1999; 19: 1049–1055.
Hong H, Kohli K, Garabedian MJ, Stallcup MR . GRIP1, a transcriptional coactivator for the AF-2 transactivation domain of steroid, thyroid, retinoid, and vitamin D receptors. Mol Cell Biol 1997; 17: 2735–2744.
Aguilera O, Pena C, Garcia JM, Larriba MJ, Ordonez-Moran P, Navarro D et al. The Wnt antagonist DICKKOPF-1 gene is induced by 1alpha,25-dihydroxyvitamin D3 associated to the differentiation of human colon cancer cells. Carcinogenesis 2007; 28: 1877–1884.
Mulholland DJ, Dedhar S, Coetzee GA, Nelson CC . Interaction of nuclear receptors with the Wnt/beta-catenin/Tcf signaling axis: Wnt you like to know? Endocr Rev 2005; 26: 898–915.
Easwaran V, Pishvaian M, Salimuddin, Byers S . Cross-regulation of beta-catenin-LEF/TCF and retinoid signaling pathways. Curr Biol 1999; 9: 1415–1418.
Kefalopoulou Z, Tzelepi V, Zolota V, Grivas PD, Christopoulos C, Kalofonos H et al. Prognostic value of novel biomarkers in astrocytic brain tumors: nuclear receptor co-regulators AIB1, TIF2, and PELP1 are associated with high tumor grade and worse patient prognosis. J Neurooncol 2012; 106: 23–31.
Taylor BS, Schultz N, Hieronymus H, Gopalan A, Xiao Y, Carver BS et al. Integrative genomic profiling of human prostate cancer. Cancer Cell 2010; 18: 11–22.
Malkova A, Ira G . Break-induced replication: functions and molecular mechanism. Curr Opin Genet Dev 2013; 23: 271–279.
Maher CA, Wilson RK . Chromothripsis and human disease: piecing together the shattering process. Cell 2012; 148: 29–32.
Walfish PG, Yoganathan T, Yang YF, Hong H, Butt TR, Stallcup MR . Yeast hormone response element assays detect and characterize GRIP1 coactivator-dependent activation of transcription by thyroid and retinoid nuclear receptors. Proc Natl Acad Sci USA 1997; 94: 3697–3702.
Voegel JJ, Heine MJ, Zechel C, Chambon P, Gronemeyer H . TIF2, a 160 kDa transcriptional mediator for the ligand-dependent activation function AF-2 of nuclear receptors. EMBO J 1996; 15: 3667–3675.
Hong H, Kohli K, Trivedi A, Johnson DL, Stallcup MR . GRIP1, a novel mouse protein that serves as a transcriptional coactivator in yeast for the hormone binding domains of steroid receptors. Proc Natl Acad Sci USA 1996; 93: 4948–4952.
D'Errico I, Moschetta A . Nuclear receptors, intestinal architecture and colon cancer: an intriguing link. Cell Mol Life Sci 2008; 65: 1523–1543.
Gingeras TR . Implications of chimaeric non-co-linear transcripts. Nature 2009; 461: 206–211.
Akiva P, Toporik A, Edelheit S, Peretz Y, Diber A, Shemesh R et al. Transcription-mediated gene fusion in the human genome. Genome Res 2006; 16: 30–36.
Herai RH, Yamagishi ME . Detection of human interchromosomal trans-splicing in sequence databanks. Brief Bioinform 2010; 11: 198–209.
Maher CA, Palanisamy N, Brenner JC, Cao X, Kalyana-Sundaram S, Luo S et al. Chimeric transcript discovery by paired-end transcriptome sequencing. Proc Natl Acad Sci USA 2009; 106: 12353–12358.
Xi Y, Formentini A, Chien M, Weir DB, Russo JJ, Ju J et al. Prognostic values of microRNAs in colorectal cancer. Biomarker Insights 2006; 2: 113–121.
Wu J, Ji X, Zhu L, Jiang Q, Wen Z, Xu S et al. Up-regulation of microRNA-1290 impairs cytokinesis and affects the reprogramming of colon cancer cells. Cancer Lett 2013; 329: 155–163.
Blackledge NP, Zhou JC, Tolstorukov MY, Farcas AM, Park PJ, Klose RJ . CpG islands recruit a histone H3 lysine 36 demethylase. Mol Cell 2010; 38: 179–190.
Frenkel-Morgenstern M, Lacroix V, Ezkurdia I, Levin Y, Gabashvili A, Prilusky J et al. Chimeras taking shape: potential functions of proteins encoded by chimeric RNA transcripts. Genome Res 2012; 22: 1231–1242.
Nothwang HG, Kim HG, Aoki J, Geisterfer M, Kubart S, Wegner RD et al. Functional hemizygosity of PAFAH1B3 due to a PAFAH1B3-CLK2 fusion gene in a female with mental retardation, ataxia and atrophy of the brain. Hum Mol Genet 2001; 10: 797–806.
Yoshida H, Miyachi M, Sakamoto K, Ouchi K, Yagyu S, Kikuchi K et al. PAX3-NCOA2 fusion gene has a dual role in promoting the proliferation and inhibiting the myogenic differentiation of rhabdomyosarcoma cells. Oncogene 2014; 33: 5601–5608.
Nakayama R, Miura Y, Ogino J, Susa M, Watanabe I, Horiuchi K et al. Detection of HEY1-NCOA2 fusion by fluorescence in-situ hybridization in formalin-fixed paraffin-embedded tissues as a possible diagnostic tool for mesenchymal chondrosarcoma. Pathol Int 2012; 62: 823–826.
Arbajian E, Magnusson L, Mertens F, Domanski HA, Vult von Steyern F, Nord KH . A novel GTF2I/NCOA2 fusion gene emphasizes the role of NCOA2 in soft tissue angiofibroma development. Genes Chromosomes Cancer 2013; 52: 330–331.
Strehl S, Nebral K, Konig M, Harbott J, Strobl H, Ratei R et al. ETV6-NCOA2: a novel fusion gene in acute leukemia associated with coexpression of T-lymphoid and myeloid markers and frequent NOTCH1 mutations. Clin Cancer Res 2008; 14: 977–983.
Liang J, Prouty L, Williams BJ, Dayton MA, Blanchard KL . Acute mixed lineage leukemia with an inv(8)(p11q13) resulting in fusion of the genes for MOZ and TIF2. Blood 1998; 92: 2118–2122.
Carapeti M, Aguiar RC, Goldman JM, Cross NC . A novel fusion between MOZ and the nuclear receptor coactivator TIF2 in acute myeloid leukemia. Blood 1998; 91: 3127–3133.
Acknowledgements
Grant Support: The work was supported by Shenzhen Municipal Science and Technology R&D fund (JCYJ20120619152326450), the Shenzhen Technology and Innovation Project Fund (JSGG20130412171021059), 863 Program China (2012AA02A506), 973 Program China (2013CB531401), the Theme-based Research Scheme of the Hong Kong Research Grants Council (T12-403-11) and the Shenzhen Virtual University Park Support Scheme to CUHK Shenzhen Research Institute.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Oncogene website
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/4.0/
About this article
Cite this article
Yu, J., Wu, W., Liang, Q. et al. Disruption of NCOA2 by recurrent fusion with LACTB2 in colorectal cancer. Oncogene 35, 187–195 (2016). https://doi.org/10.1038/onc.2015.72
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/onc.2015.72
This article is cited by
-
Identification of the prognostic value of LACTB2 and its correlation with immune infiltrates in ovarian cancer by integrated bioinformatics analyses
European Journal of Medical Research (2024)
-
Proteome-wide identification of arginine methylation in colorectal cancer tissues from patients
Proteome Science (2020)
-
Targeting epigenetic regulators for cancer therapy: mechanisms and advances in clinical trials
Signal Transduction and Targeted Therapy (2019)
-
Mesenchymal Chondrosarcoma: a Review with Emphasis on its Fusion-Driven Biology
Current Oncology Reports (2018)
-
Involvement of DPP9 in gene fusions in serous ovarian carcinoma
BMC Cancer (2017)