Abstract
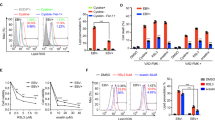
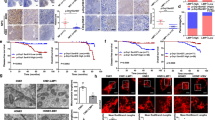
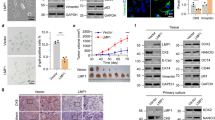
Epstein-Barr virus (EBV) causes human lymphoid malignancies, and the EBV product latent membrane protein 1 (LMP1) has been identified as an oncogene in epithelial carcinomas such as nasopharyngeal carcinoma (NPC). EBV can epigenetically reprogram lymphocyte-specific processes and induce cell immortalization. However, the interplay between LMP1 and the NPC host cell remains largely unknown. Here, we report that LMP1 is important to establish the Hox gene expression signature in NPC cell lines and tumor biopsies. LMP1 induces repression of several Hox genes in part via stalling of RNA polymerase II (RNA Pol II). Pol II stalling can be overcome by irradiation involving the epigenetic regulator TET3. Furthermore, we report that HoxC8, one of the genes silenced by LMP1, has a role in tumor growth. Ectopic expression of HoxC8 inhibits NPC cell growth in vitro and in vivo, modulates glycolysis and regulates the expression of tricarboxylic acid (TCA) cycle-related genes. We propose that viral latency products may repress via stalling key mediators that in turn modulate glycolysis.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
References
Levine M . Paused RNA polymerase II as a developmental checkpoint. Cell 2011; 145: 502–511.
Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z et al. High-resolution profiling of histone methylations in the human genome. Cell 2007; 129: 823–837.
Core LJ, Waterfall JJ, Lis JT . Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science 2008; 322: 1845–1848.
Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA . A chromatin landmark and transcription initiation at most promoters in human cells. Cell 2007; 130: 77–88.
Muse GW, Gilchrist DA, Nechaev S, Shah R, Parker JS, Grissom SF et al. RNA polymerase is poised for activation across the genome. Nat Genet 2007; 39: 1507–1511.
Liu S, Tao Y . Interplay between chromatin modifications and paused RNA polymerase II in dynamic transition between stalled and activated genes. Biol Rev Camb Philos Soc 2013; 88: 40–48.
Feng S, Cokus SJ, Zhang X, Chen PY, Bostick M, Goll MG et al. Conservation and divergence of methylation patterning in plants and animals. Proc Natl Acad Sci USA 2010; 107: 8689–8694.
Tanay A, O'Donnell AH, Damelin M, Bestor TH . Hyperconserved CpG domains underlie Polycomb-binding sites. Proc Natl Acad Sci USA 2007; 104: 5521–5526.
Zemach A, McDaniel IE, Silva P, Zilberman D . Genome-wide evolutionary analysis of eukaryotic DNA methylation. Science 2010; 328: 916–919.
Mohn F, Weber M, Rebhan M, Roloff TC, Richter J, Stadler MB et al. Lineage-specific polycomb targets and de novo DNA methylation define restriction and potential of neuronal progenitors. Mol Cell 2008; 30: 755–766.
Takeshima H, Yamashita S, Shimazu T, Niwa T, Ushijima T . The presence of RNA polymerase II, active or stalled, predicts epigenetic fate of promoter CpG islands. Genome Res 2009; 19: 1974–1982.
Tao Y, Xi S, Briones V, Muegge K . Lsh mediated RNA polymerase II stalling at HoxC6 and HoxC8 involves DNA methylation. PLoS One 2010; 5: e9163.
Weber M, Hellmann I, Stadler MB, Ramos L, Paabo S, Rebhan M et al. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nat Genet 2007; 39: 457–466.
Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science 2009; 324: 930–935.
Tao Y, Liu S, Briones V, Geiman TM, Muegge K . Treatment of breast cancer cells with DNA demethylating agents leads to a release of Pol II stalling at genes with DNA-hypermethylated regions upstream of TSS. Nucleic Acids Res 2011; 39: 9508–9520.
Jiang Y, Liu S, Chen X, Cao Y, Tao Y . Genome-wide distribution of DNA methylation and DNA demethylation and related chromatin regulators in cancer. Biochim Biophys Acta 2013; 1835: 155–163.
Williams K, Christensen J, Pedersen MT, Johansen JV, Cloos PA, Rappsilber J et al. TET1 and hydroxymethylcytosine in transcription and DNA methylation fidelity. Nature 2011; 473: 343–348.
Klein E, Kis LL, Klein G . Epstein-Barr virus infection in humans: from harmless to life endangering virus-lymphocyte interactions. Oncogene 2007; 26: 1297–1305.
Plottel CS, Blaser MJ . Microbiome and malignancy. Cell Host Microbe 2011; 10: 324–335.
Palermo RD, Webb HM, West MJ . RNA polymerase II stalling promotes nucleosome occlusion and pTEFb recruitment to drive immortalization by Epstein-Barr virus. PLoS Pathog 2011; 7: e1002334.
Mesri EA, Feitelson MA, Munger K . Human viral oncogenesis: a cancer hallmarks analysis. Cell Host Microbe 2014; 15: 266–282.
Raab-Traub N . Epstein–Barr virus transforming proteins: biologic properties and contribution to oncogenesis. In Damania B, Pipas JM (eds) DNA Tumor Viruses. Springer, New York 2009, pp 259–284.
Zheng H, Li LL, Hu DS, Deng XY, Cao Y . Role of Epstein-Barr virus encoded latent membrane protein 1 in the carcinogenesis of nasopharyngeal carcinoma. Cell Mol Immunol 2007; 4: 185–196.
Lin X, Tang M, Tao Y, Li L, Liu S, Guo L et al. Epstein-Barr virus-encoded LMP1 triggers regulation of the ERK-mediated Op18/stathmin signaling pathway in association with cell cycle. Cancer Sci 2012; 103: 993–999.
Ma X, Yang L, Xiao L, Tang M, Liu L, Li Z et al. Down-regulation of EBV-LMP1 radio-sensitizes nasal pharyngeal carcinoma cells via NF-kappaB regulated ATM expression. PLoS One 2011; 6: e24647.
Niller HH, Wolf H, Minarovits J . Epigenetic dysregulation of the host cell genome in Epstein-Barr virus-associated neoplasia. Semin Cancer Biol 2009; 19: 158–164.
Skalska L, White RE, Franz M, Ruhmann M, Allday MJ . Epigenetic repression of p16(INK4A) by latent Epstein-Barr virus requires the interaction of EBNA3A and EBNA3C with CtBP. PLoS Pathog 2010; 6: e1000951.
Chopra VS, Hong JW, Levine M . Regulation of Hox gene activity by transcriptional elongation in Drosophila. Curr Biol 2009; 19: 688–693.
Shah N, Sukumar S . The Hox genes and their roles in oncogenesis. Nat Rev Cancer 2010; 10: 361–371.
Xiao L, Hu ZY, Dong X, Tan Z, Li W, Tang M et al. Targeting Epstein-Barr virus oncoprotein LMP1-mediated glycolysis sensitizes nasopharyngeal carcinoma to radiation therapy. Oncogene 2014; 33: 4568–4578.
Pogribny I, Raiche J, Slovack M, Kovalchuk O . Dose-dependence, sex- and tissue-specificity, and persistence of radiation-induced genomic DNA methylation changes. Biochem Biophys Res Commun 2004; 320: 1253–1261.
DeBerardinis RJ, Thompson CB . Cellular metabolism and disease: what do metabolic outliers teach us? Cell 2012; 148: 1132–1144.
Chopra VS, Cande J, Hong JW, Levine M . Stalled Hox promoters as chromosomal boundaries. Genes Dev 2009; 23: 1505–1509.
Bijl J, Krosl J, Lebert-Ghali CE, Vacher J, Mayotte N, Sauvageau G . Evidence for Hox and E2A-PBX1 collaboration in mouse T-cell leukemia. Oncogene 2008; 27: 6356–6364.
Hyland PL, McDade SS, McCloskey R, Dickson GJ, Arthur K, McCance DJ et al. Evidence for alteration of EZH2, BMI1, and KDM6A and epigenetic reprogramming in human papillomavirus type 16 E6/E7-expressing keratinocytes. J Virol 2011; 85: 10999–11006.
McLaughlin-Drubin ME, Crum CP, Munger K . Human papillomavirus E7 oncoprotein induces KDM6A and KDM6B histone demethylase expression and causes epigenetic reprogramming. Proc Natl Acad Sci USA 2011; 108: 2130–2135.
Rodini CO, Xavier FC, Paiva KB, De Souza Setubal Destro MF, Moyses RA, Michaluarte P et al. Homeobox gene expression profile indicates HOXA5 as a candidate prognostic marker in oral squamous cell carcinoma. Int J Oncol 2012; 40: 1180–1188.
Marra L, Cantile M, Scognamiglio G, Perdona S, La Mantia E, Cerrone M et al. Deregulation of HOX B13 expression in urinary bladder cancer progression. Curr Med Chem 2013; 20: 833–839.
Chen J, Zhu S, Jiang N, Shang Z, Quan C, Niu Y . HoxB3 promotes prostate cancer cell progression by transactivating CDCA3. Cancer Lett 2013; 330: 217–224.
Adwan H, Zhivkova-Galunska M, Georges R, Eyol E, Kleeff J, Giese NA et al. Expression of HOXC8 is inversely related to the progression and metastasis of pancreatic ductal adenocarcinoma. Br J Cancer 2011; 105: 288–295.
Du YB, Dong B, Shen LY, Yan WP, Dai L, Xiong HC et al. The survival predictive significance of HOXC6 and HOXC8 in esophageal squamous cell carcinoma. J Surg Res 2014; 188: 442–450.
Li Y, Chao F, Huang B, Liu D, Kim J, Huang S . HOXC8 promotes breast tumorigenesis by transcriptionally facilitating cadherin-11 expression. Oncotarget 2014; 5: 2596–2607.
Carmona FJ, Villanueva A, Vidal A, Munoz C, Puertas S, Penin RM et al. Epigenetic disruption of cadherin-11 in human cancer metastasis. J Pathol 2012; 228: 230–240.
Li L, Ying J, Li H, Zhang Y, Shu X, Fan Y et al. The human cadherin 11 is a pro-apoptotic tumor suppressor modulating cell stemness through Wnt/beta-catenin signaling and silenced in common carcinomas. Oncogene 2012; 31: 3901–3912.
Uchida J, Yasui T, Takaoka-Shichijo Y, Muraoka M, Kulwichit W, Raab-Traub N et al. Mimicry of CD40 signals by Epstein-Barr virus LMP1 in B lymphocyte responses. Science 1999; 286: 300–303.
Verweij FJ, van Eijndhoven MA, Hopmans ES, Vendrig T, Wurdinger T, Cahir-McFarland E et al. LMP1 association with CD63 in endosomes and secretion via exosomes limits constitutive NF-kappaB activation. EMBO J 2011; 30: 2115–2129.
Xu J, Ahmad A, Menezes J . Preferential localization of the Epstein-Barr virus (EBV) oncoprotein LMP-1 to nuclei in human T cells: implications for its role in the development of EBV genome-positive T-cell lymphomas. J Virol 2002; 76: 4080–4086.
Hau PM, Tsang CM, Yip YL, Huen MS, Tsao SW . Id1 interacts and stabilizes the Epstein-Barr virus latent membrane protein 1 (LMP1) in nasopharyngeal epithelial cells. PLoS One 2011; 6: e21176.
Lai W, Li H, Liu S, Tao Y . Connecting chromatin modifying factors to DNA damage response. Int J Mol Sci 2013; 14: 2355–2369.
Liu S, Tao Y, Chen X, Cao Y . The dynamic interplay in chromatin remodeling factors polycomb and trithorax proteins in response to DNA damage. Mol Biol Rep 2012; 39: 6179–6185.
Tusher VG, Tibshirani R, Chu G . Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA 2001; 98: 5116–5121.
Cortazar D, Kunz C, Selfridge J, Lettieri T, Saito Y, MacDougall E et al. Embryonic lethal phenotype reveals a function of TDG in maintaining epigenetic stability. Nature 2011; 470: 419–423.
Cortellino S, Xu J, Sannai M, Moore R, Caretti E, Cigliano A et al. Thymine DNA glycosylase is essential for active DNA demethylation by linked deamination-base excision repair. Cell 2011; 146: 67–79.
Hajkova P, Jeffries SJ, Lee C, Miller N, Jackson SP, Surani MA . Genome-wide reprogramming in the mouse germ line entails the base excision repair pathway. Science 2010; 329: 78–82.
Popp C, Dean W, Feng S, Cokus SJ, Andrews S, Pellegrini M et al. Genome-wide erasure of DNA methylation in mouse primordial germ cells is affected by AID deficiency. Nature 2010; 463: 1101–1105.
Misteli T, Soutoglou E . The emerging role of nuclear architecture in DNA repair and genome maintenance. Nat Rev Mol Cell Biol 2009; 10: 243–254.
Merrifield M, Kovalchuk O . Epigenetics in radiation biology: a new research frontier. Front Genet 2013; 4: 40.
Smits KM, Melotte V, Niessen HE, Dubois L, Oberije C, Troost EG et al. Epigenetics in radiotherapy: where are we heading? Radiotherapy and oncology 2014; 111: 168–177.
Pastor WA, Aravind L, Rao A . TETonic shift: biological roles of TET proteins in DNA demethylation and transcription. Nat Rev Mol Cell Biol 2013; 14: 341–356.
Lian CG, Xu Y, Ceol C, Wu F, Larson A, Dresser K et al. Loss of 5-hydroxymethylcytosine is an epigenetic hallmark of melanoma. Cell 2012; 150: 1135–1146.
Song SJ, Poliseno L, Song MS, Ala U, Webster K, Ng C et al. MicroRNA-antagonism regulates breast cancer stemness and metastasis via TET-family-dependent chromatin remodeling. Cell 2013; 154: 311–324.
Yang H, Liu Y, Bai F, Zhang JY, Ma SH, Liu J et al. Tumor development is associated with decrease of TET gene expression and 5-methylcytosine hydroxylation. Oncogene 2013; 32: 663–669.
Cairns RA, Harris IS, Mak TW . Regulation of cancer cell metabolism. Nat Rev Cancer 2011; 11: 85–95.
Galluzzi L, Kepp O, Vander Heiden MG, Kroemer G . Metabolic targets for cancer therapy. Nat Rev Drug Discov 2013; 12: 829–846.
Oermann EK, Wu J, Guan KL, Xiong Y . Alterations of metabolic genes and metabolites in cancer. Semin Cell Dev Biol 2012; 23: 370–380.
Xiao M, Yang H, Xu W, Ma S, Lin H, Zhu H et al. Inhibition of alpha-KG-dependent histone and DNA demethylases by fumarate and succinate that are accumulated in mutations of FH and SDH tumor suppressors. Genes Dev 2012; 26: 1326–1338.
Poteet E, Choudhury GR, Winters A, Li W, Ryou MG, Liu R et al. Reversing the Warburg effect as a treatment for glioblastoma. J Biol Chem 2013; 288: 9153–9164.
Sancak Y, Peterson TR, Shaul YD, Lindquist RA, Thoreen CC, Bar-Peled L et al. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science 2008; 320: 1496–1501.
Shi Y, Tao Y, Jiang Y, Xu Y, Yan B, Chen X et al. Nuclear epidermal growth factor receptor interacts with transcriptional intermediary factor 2 to activate cyclin D1 gene expression triggered by the oncoprotein latent membrane protein 1. Carcinogenesis 2012; 33: 1468–1478.
Olive PL, Banath JP . The comet assay: a method to measure DNA damage in individual cells. Nat Protoc 2006; 1: 23–29.
Acknowledgements
This work was supported by the National Basic Research Program of China (2011CB504300 (YT); 2015CB553903 (YT)); the Hunan Natural Science Foundation of China (12JJ1013 (YT)); the Fundamental Research Funds for the Central Universities (2011JQ019 (YT), 2013ZZTS074 (BY), 2013ZZTS284 (WL)); and the National Natural Science Foundation of China (81171881 and 81372427 (YT), 81271763 (SL), 81302354 (YS)); and the Hunan Provincial Innovation Foundation For Postgraduate (71380100002 (YJ)). This project has been funded in part with Federal funds from the Frederick National Laboratory for Cancer Research, National Institutes of Health, under contract HHSN261200800001E. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products or organizations imply endorsement by the US Government.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Oncogene website
Supplementary information
Rights and permissions
About this article
Cite this article
Jiang, Y., Yan, B., Lai, W. et al. Repression of Hox genes by LMP1 in nasopharyngeal carcinoma and modulation of glycolytic pathway genes by HoxC8. Oncogene 34, 6079–6091 (2015). https://doi.org/10.1038/onc.2015.53
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/onc.2015.53
This article is cited by
-
Targeting the signaling in Epstein–Barr virus-associated diseases: mechanism, regulation, and clinical study
Signal Transduction and Targeted Therapy (2021)
-
Annotation and cluster analysis of long noncoding RNA linked to male sex and estrogen in cancers
npj Precision Oncology (2020)
-
Bmi1 drives hepatocarcinogenesis by repressing the TGFβ2/SMAD signalling axis
Oncogene (2020)
-
The deubiquitylase UCHL3 maintains cancer stem-like properties by stabilizing the aryl hydrocarbon receptor
Signal Transduction and Targeted Therapy (2020)
-
Viral hijacking of cellular metabolism
BMC Biology (2019)