Abstract
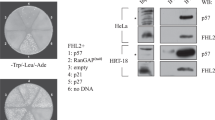
KDM2B (also known as FBXL10) controls stem cell self-renewal, somatic cell reprogramming and senescence, and tumorigenesis. KDM2B contains multiple functional domains, including a JmjC domain that catalyzes H3K36 demethylation and a CxxC zinc-finger that recognizes CpG islands and recruits the polycomb repressive complex 1. Here, we report that KDM2B, via its F-box domain, functions as a subunit of the CUL1-RING ubiquitin ligase (CRL1/SCFKDM2B) complex. KDM2B targets c-Fos for polyubiquitylation and regulates c-Fos protein levels. Unlike the phosphorylation of other SCF (SKP1-CUL1-F-box)/CRL1 substrates that promotes substrates binding to F-box, epidermal growth factor (EGF)-induced c-Fos S374 phosphorylation dissociates c-Fos from KDM2B and stabilizes c-Fos protein. Non-phosphorylatable and phosphomimetic mutations at S374 result in c-Fos protein which cannot be induced by EGF or accumulates constitutively and lead to decreased or increased cell proliferation, respectively. Multiple tumor-derived KDM2B mutations impaired the function of KDM2B to target c-Fos degradation and to suppress cell proliferation. These results reveal a novel function of KDM2B in the negative regulation of cell proliferation by assembling an E3 ligase to targeting c-Fos protein degradation that is antagonized by mitogenic stimulations.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Eferl R, Wagner EF . AP-1: a double-edged sword in tumorigenesis. Nat Rev Cancer 2003; 3: 859–868.
Shaulian E, Karin M . AP-1 as a regulator of cell life and death. Nat Cell Biol 2002; 4: E131–E136.
Greenberg ME, Ziff EB . Stimulation of 3T3 cells induces transcription of the c-fos proto-oncogene. Nature 1984; 311: 433–438.
Rauscher FJ III, Sambucetti LC, Curran T, Distel RJ, Spiegelman BM . Common DNA binding site for Fos protein complexes and transcription factor AP-1. Cell 1988; 52: 471–480.
Chiu R, Boyle WJ, Meek J, Smeal T, Hunter T, Karin M . The c-Fos protein interacts with c-Jun/AP-1 to stimulate transcription of AP-1 responsive genes. Cell 1988; 54: 541–552.
Sassone-Corsi P, Ransone LJ, Lamph WW, Verma IM . Direct interaction between fos and jun nuclear oncoproteins: role of the 'leucine zipper' domain. Nature 1988; 336: 692–695.
Glover JN, Harrison SC . Crystal structure of the heterodimeric bZIP transcription factor c-Fos-c-Jun bound to DNA. Nature 1995; 373: 257–261.
Nakakuki T, Birtwistle MR, Saeki Y, Yumoto N, Ide K, Nagashima T et al. Ligand-specific c-Fos expression emerges from the spatiotemporal control of ErbB network dynamics. Cell 2010; 141: 884–896.
Treisman R . Journey to the surface of the cell: Fos regulation and the SRE. EMBO J 1995; 14: 4905–4913.
Gille H, Sharrocks AD, Shaw PE . Phosphorylation of transcription factor p62TCF by MAP kinase stimulates ternary complex formation at c-fos promoter. Nature 1992; 358: 414–417.
Okazaki K, Sagata N . The Mos/MAP kinase pathway stabilizes c-Fos by phosphorylation and augments its transforming activity in NIH 3T3 cells. EMBO J 1995; 14: 5048–5059.
Stancovski I, Gonen H, Orian A, Schwartz AL, Ciechanover A . Degradation of the proto-oncogene product c-Fos by the ubiquitin proteolytic system in vivo and in vitro: identification and characterization of the conjugating enzymes. Mol Cell Biol 1995; 15: 7106–7116.
Chen RH, Abate C, Blenis J . Phosphorylation of the c-Fos transrepression domain by mitogen-activated protein kinase and 90-kDa ribosomal S6 kinase. Proc Natl Acad Sci USA 1993; 90: 10952–10956.
Bakiri L, Reschke MO, Gefroh HA, Idarraga MH, Polzer K, Zenz R et al. Functions of Fos phosphorylation in bone homeostasis, cytokine response and tumourigenesis. Oncogene 2011; 30: 1506–1517.
Sasaki T, Kojima H, Kishimoto R, Ikeda A, Kunimoto H, Nakajima K . Spatiotemporal regulation of c-Fos by ERK5 and the E3 ubiquitin ligase UBR1, and its biological role. Mol Cell 2006; 24: 63–75.
Gilley R, March HN, Cook SJ . ERK1/2, but not ERK5, is necessary and sufficient for phosphorylation and activation of c-Fos. Cell Signal 2009; 21: 969–977.
He J, Shen L, Wan M, Taranova O, Wu H, Zhang Y . Kdm2b maintains murine embryonic stem cell status by recruiting PRC1 complex to CpG islands of developmental genes. Nat Cell Biol 2013; 15: 373–384.
Liang G, He J, Zhang Y . Kdm2b promotes induced pluripotent stem cell generation by facilitating gene activation early in reprogramming. Nat Cell Biol 2012; 14: 457–466.
He J, Kallin EM, Tsukada Y, Zhang Y . The H3K36 demethylase Jhdm1b/Kdm2b regulates cell proliferation and senescence through p15(Ink4b). Nat Struct Mol Biol 2008; 15: 1169–1175.
Pfau R, Tzatsos A, Kampranis SC, Serebrennikova OB, Bear SE, Tsichlis PN . Members of a family of JmjC domain-containing oncoproteins immortalize embryonic fibroblasts via a JmjC domain-dependent process. Proc Natl Acad Sci USA 2008; 105: 1907–1912.
Kottakis F, Foltopoulou P, Sanidas I, Keller P, Wronski A, Dake BT et al. NDY1/KDM2B functions as a master regulator of polycomb complexes and controls self-renewal of breast cancer stem cells. Cancer Res 2014; 74: 3935–3946.
He J, Nguyen AT, Zhang Y . KDM2b/JHDM1b, an H3K36me2-specific demethylase, is required for initiation and maintenance of acute myeloid leukemia. Blood 2011; 117: 3869–3880.
Tzatsos A, Paskaleva P, Ferrari F, Deshpande V, Stoykova S, Contino G et al. KDM2B promotes pancreatic cancer via Polycomb-dependent and -independent transcriptional programs. J Clin Invest 2013; 123: 727–739.
Tsukada Y, Fang J, Erdjument-Bromage H, Warren ME, Borchers CH, Tempst P et al. Histone demethylation by a family of JmjC domain-containing proteins. Nature 2006; 439: 811–816.
Farcas AM, Blackledge NP, Sudbery I, Long HK, McGouran JF, Rose NR et al. KDM2B links the polycomb repressive complex 1 (PRC1) to recognition of CpG islands. eLife 2012; 1: e00205.
Wu X, Johansen Jens V, Helin K . Fbxl10/Kdm2b recruits polycomb repressive complex 1 to CpG islands and regulates H2A ubiquitylation. Mol Cell 2013; 49: 1134–1146.
Gearhart MD, Corcoran CM, Wamstad JA, Bardwell VJ . Polycomb group and SCF ubiquitin ligases are found in a novel BCOR complex that is recruited to BCL6 targets. Mol Cell Biol 2006; 26: 6880–6889.
Koyama-Nasu R, David G, Tanese N . The F-box protein Fbl10 is a novel transcriptional repressor of c-Jun. Nat Cell Biol 2007; 9: 1074–1080.
Bai C, Sen P, Hofmann K, Ma L, Goebl M, Harper JW et al. SKP1 connects cell cycle regulators to the ubiquitin proteolysis machinery through a novel motif, the F-box. Cell 1996; 86: 263–274.
Skowyra D, Craig KL, Tyers M, Elledge SJ, Harper JW . F-box proteins are receptors that recruit phosphorylated substrates to the SCF ubiquitin-ligase complex. Cell 1997; 91: 209–219.
Feldman RM, Correll CC, Kaplan KB, Deshaies RJ . A complex of Cdc4p, Skp1p, and Cdc53p/cullin catalyzes ubiquitination of the phosphorylated CDK inhibitor Sic1p. Cell 1997; 91: 221–230.
Zheng N, Schulman BA, Song L, Miller JJ, Jeffrey PD, Wang P et al. Structure of the Cul1-Rbx1-Skp1-F boxSkp2 SCF ubiquitin ligase complex. Nature 2002; 416: 703–709.
Bartke T, Vermeulen M, Xhemalce B, Robson SC, Mann M, Kouzarides T . Nucleosome-interacting proteins regulated by DNA and histone methylation. Cell 2010; 143: 470–484.
Maher VM, Heflich RH, McCormick JJ . Repair of DNA damage induced in human fibroblasts by N-substituted aryl compounds. Natl Cancer Inst Monogr 1981; 58: 217–222.
Furukawa M, Andrews PS, Xiong Y . Assays for RING family ubiquitin ligases. Methods Mol Biol 2005; 301: 37–46.
Acknowledgements
We thank the members of the Fudan MCB laboratory for discussions and support throughout this study and Michele Pagano of NYU for providing plasmids expressing full-length human KDM2B cDNA. This work was supported by Chinese Ministry of Sciences and Technology 973 (Grant No. 2015CB910401), NSFC (Grant No. 81225016, 81430057), Shanghai Key basic research program (12JC1401100), Shanghai Outstanding Academic Leader (Grant No.13XD1400600) and the Youth Science and Technology Leading Talent by MOST (to Q-YL), NIH Grants EY022611 and CA132809 (to K-LG) and GM067113 (to YX).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Oncogene website
Supplementary information
Rights and permissions
About this article
Cite this article
Han, XR., Zha, Z., Yuan, HX. et al. KDM2B/FBXL10 targets c-Fos for ubiquitylation and degradation in response to mitogenic stimulation. Oncogene 35, 4179–4190 (2016). https://doi.org/10.1038/onc.2015.482
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/onc.2015.482
This article is cited by
-
The emerging roles of histone demethylases in cancers
Cancer and Metastasis Reviews (2024)
-
FBXL10 promotes EMT and metastasis of breast cancer cells via regulating the acetylation and transcriptional activity of SNAI1
Cell Death Discovery (2021)
-
KDM2B promotes IL-6 production and inflammatory responses through Brg1-mediated chromatin remodeling
Cellular & Molecular Immunology (2020)
-
miR-146b promotes cell proliferation and increases chemosensitivity, but attenuates cell migration and invasion via FBXL10 in ovarian cancer
Cell Death & Disease (2018)