Abstract
Inositol polyphosphate 4-phosphatase type II (INPP4B) negatively regulates phosphatidylinositol 3-kinase signaling and is a tumor suppressor in some types of cancers. However, we have found that it is frequently upregulated in human colon cancer cells. Here we show that silencing of INPP4B blocks activation of Akt and serum- and glucocorticoid-regulated kinase 3 (SGK3), inhibits colon cancer cell proliferation and retards colon cancer xenograft growth. Conversely, overexpression of INPP4B increases proliferation and triggers anchorage-independent growth of normal colon epithelial cells. Moreover, we demonstrate that the effect of INPP4B on Akt and SGK3 is associated with inactivation of phosphate and tensin homolog through its protein phosphatase activity and that the increase in INPP4B is due to Ets-1-mediated transcriptional upregulation in colon cancer cells. Collectively, these results suggest that INPP4B may function as an oncogenic driver in colon cancer, with potential implications for targeting INPP4B as a novel approach to treat this disease.
Similar content being viewed by others
Introduction
Aberrant activation of survival-signaling pathways has an important role in cancer development, progression and resistance to treatment.1, 2 In colon cancer (CRC), activation of the phosphatidylinositol 3-kinase (PI3K) pathway is of particular importance, in that many common genetic and epigenetic anomalies in the disease, such as amplification of epidermal growth factor receptor, activating mutations of KRAS and loss of phosphate and tensin homolog deleted on chromosome 10 (PTEN), converge on activation of PI3K signaling.3
PI3K signaling is initiated with the engagement of extracellular growth factors to receptor tyrosine kinases. This results in recruitment of PI3K to plasma membrane-anchored receptors where it is activated, leading to increases in the production of phosphatidylinositol(3,4) bisphosphate (PI(3,4)P2) and phosphatidylinositol(3,4,5) trisphosphate (PI(3,4,5)P3), which in turn bind to and activate multiple downstream effectors.4, 5 Among them is Akt, which, upon activation, phosphorylates a large array of substrates to promote cell survival and proliferation and contributes to the pathogenesis of cancer.
PI3K can also drive oncogenic signaling independently of Akt through activation of serum- and glucocorticoid-regulated kinase (SGK), another family of serine/threonine kinases consisting of three isoforms, SGK1, SGK2 and SGK3,6 which are highly homologous to and share substrate specificity with Akt. Among SGK isoforms, SGK3 is unique in that it contains an N-terminal PX domain that enables its binding to PI(3)P, thus targeting it to early endosomes where it is fully activated.6, 7
Activation of PI3K signaling is negatively regulated by three classes of inositol polyphosphate phosphatases.8, 9, 10 The inositol polyphosphate 3-phosphatase (3-phosphatase) PTEN dephosphorylates the 3-position of PI(3,4,5)P3 to generate PI(4,5)P2 (refs. 11, 12), whereas 5-phosphatases, such as Src homology 2-containing inositol 5- phosphatase (SHIP) and phosphatidylinositol 4,5-bisphosphate 5-phosphatase (PIB5PA)/proline-rich inositol polyphosphate phosphatase (PIPP) dephosphorylate the 5-position to produce PI(3,4)P2.13, 14 The latter is in turn subjected to dephosphorylation by inositol polyphosphate 4-phosphatase type I (INPP4A) and type II (INPP4B) at the 4-position to generate PI(3)P, thus terminating PI3K signaling.9, 15
Although PTEN is a well-established tumor suppressor,11, 12 some 5-phosphatases such as SHIP2 and PIB5PA and the 4-phosphatase INPP4B are also tumor suppressive through inhibition of PI3K signaling in a variety of types of cancers.8, 9, 13 Nevertheless, INPP4B-dependent activation of SGK3 drives tumourigenesis in a subset of breast cancers with low Akt.16 Moreover, INPP4B is associated with chemoresistance and poor outcome of patients with acute myeloid leukemia.17, 18 Here we report that INPP4B is frequently upregulated in human colon cancers and has an important role in colon cancer cell proliferation. In addition, we show that the effect of INPP4B on colon cancer is mediated by activation of Akt and SGK3 that is associated with downregulation of PTEN through its protein phosphatase activity and that upregulation of INPP4B is due to an Ets-1-mediated transcriptional increase in colon cancer cells.
Results
INPP4B is frequently upregulated in human colon cancer
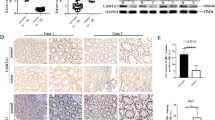
We examined the expression of INPP4B by immunohistochemistry in tissue microarrays constructed from 124 formalin-fixed paraffin-embedded colon cancers and paired adjacent noncancerous colon tissues (Supplementary Table S1). The results revealed that INPP4B expression was elevated in 82 colon cancers (66%). The average increase was 8.2 times (Figures 1a and b). However, there was no significant difference in INPP4B levels among colon cancers of different clinicopathological groups defined by stage and grade of the tumors as well as gender and age of the patients (Supplementary Table S1).
INPP4B is frequently upregulated in colon cancer cells. (a) Representative microphotographs of immunohistochemical (IHC) staining of INPP4B in tissue microarrays constructed from colon cancers and paired adjacent noncancerous colon tissues. Scale bar, 100 μm. (b) Comparison of INPP4B expression in the 124 colon cancers and paired adjacent noncancerous colon tissues as detailed in Supplementary Table S1. Data are represented as mean immunoreactive score±s.e.m. of IHC staining. ***P<0.001, Kruskal–Wallis test. (c) Comparison of INPP4B mRNA levels of laser capture micro-dissected (LCM) colon cancer cells from the 120 colon cancers and paired adjacent noncancerous epithelial tissues as detailed in Supplementary Table S2. The relative abundance of INPP4B mRNA in the latter was arbitrarily designated as 1. Data are represented as mean±s.e.m. **P<0.01, Student's t-test. (d) Comparison of INPP4B mRNA levels in LCM colon cancer cells (LCM) (n=77) and homogenized colon cancer tissues (HC) (n=17) from the Matsuyama microarray gene expression data set (http://r2.amc.nl). *P<0.05, Student's t-test. (e) Whole-cell lysates from the FHC normal colon epithelial cell line and the indicated colon cancer cell lines with different mutational status of PIK3CA, KRAS and PTEN were subjected to western blotting analysis of INPP4B and GAPDH (as a loading control). Data are representative of three individual experiments. W: wild-type; M: mutant. (f) Total RNA from the FHC normal colon epithelial cell line and the indicated colon cancer cell lines were subjected to qPCR analysis of INPP4B mRNA expression. The relative abundance of INPP4B mRNA in FHC cells was arbitrarily designated as 1. Data are represented as mean±s.e.m. of three individual experiments.
The expression of INPP4B mRNA was quantitated by quantitative PCR (qPCR) analysis of laser capture micro-dissected CRC cells from another cohort of 120 freshly removed colon cancers in comparison with paired adjacent noncancerous epithelial tissues (Supplementary Table S2). The INPP4B transcript was similarly increased in dissected CRC cells from 70% of the colon cancers with an average increase of 8.6-folds (Figure 1c and Supplementary Figure S1). In support, analysis of the publically available Matsuyama microarray gene expression data set indicated that the INPP4B mRNA expression was higher in laser capture micro-dissected CRC cells compared with homogenized colon cancer tissues that were a mixture of CRC cells and adjacent noncancerous cells (Figure 1d; http://r2.amc.nl).
We also examined the expression of INPP4B in a panel of colon cancer cell lines by immunoblotting. These colon cancer cell lines had varying status of the most common mutations of the key components of the PI3K pathway, KRAS and PIK3CA, but all carried wild-type PTEN (Figure 1e and Supplementary Table S3). None of the colon cancer cell lines harbored INPP4B mutations as shown by sequencing of all the 27 exons (including the intron/exon boundaries) of the gene. Although the INPP4B protein was readily detected in all but two colon cancer cell lines (SW480 and SW620) at various levels, it was not measurable in FHC cells (Figure 1e). Similarly, INPP4B mRNA was also increased in all but the two colon cancer cell lines compared with the normal colon epithelial cell line FHC (Figure 1f).
INPP4B promotes proliferation of colon cancer cells
We focused on examination of the functional significance of INPP4B upregulation in colon cancer cells. Strikingly, INPP4B knockdown caused reduction in the basal levels of activation of Akt and inhibited Akt activation in response to stimulation with epidermal growth factor in all the colon cancer cell lines tested (WiDr, HCT116, Lim1215, EB) regardless of their genetic backgrounds (Figures 1e, 2a and b; Supplementary Figures S2A and B; and Supplementary Table S3). Although INPP4B knockdown triggered killing of a small proportion of the cells (Supplementary Figure S2C), inhibition of cell proliferation appeared to be the predominant functional consequence (Figures 2c and d; and Supplementary Figures 2D and E). As anticipated, INPP4B knockdown enhanced Akt activation and promoted proliferation in MCF-7 cells that were used as a control (Figures 2a, c and d).8, 9 These results suggest that, despite its tumor-suppressive role in MCF-7 cells, INPP4B promotes colon cancer cell proliferation and survival, which is associated with increased activation of Akt. In support, introduction of a construct expressing shRNA-resistant cDNA of INPP4B reversed the inhibitory effect of INPP4B knockdown on cell proliferation in WiDr and HCT116 cells (Figures 2e and f). Moreover, introduction of exogenous INPP4B into SW620 cells that expressed relatively low levels of endogenous INPP4B and HT-29 cells led to increased Akt activation and cell proliferation (Figures 1e, 2g and h and Supplementary Figures S2F and G).
INPP4B promotes colon cancer cell proliferation. (a) Whole-cell lysates from WiDr and HCT116 colon cancer cells and MCF-7 breast cancer cells stably transduced with the control shRNA (shControl) or two individual INPP4B shRNAs (shINPP4B1 and shINPP4B2) were subjected to western blotting analysis of INPP4B, phosphorylated Akt (pSer473-Akt and pThr308-Akt), Akt and GAPDH (as a loading control). Data are representative of three individual experiments. (b) WiDr and HCT116 cells stably transduced with the control shRNA (shControl) or INPP4B shRNA (shINPP4B1) were treated with epidermal growth factor (100 nM) for the indicated periods. Whole-cell lysates were subjected to western blotting analysis of INPP4B, phosphorylated Akt (pSer473-Akt and pThr308-Akt), Akt, and GAPDH (as a loading control). Data are representative of three individual experiments. (c) WiDr, HCT116 and MCF-7 cells were stably transduced with the control shRNA (shControl) or two individual INPP4B shRNAs (shINPP4B1 and shINPP4B2). Forty-eight hours later, cells were subjected to bromodeoxyuridine (BrdU) incorporation assays. Data are represented as mean±s.e.m. of three individual experiments. *P<0.05, Student's t-test. (d) WiDr, HCT116 and MCF-7 cells stably transduced with the control shRNA (shControl) or two individual INPP4B shRNAs (shINPP4B1 and shINPP4B2) were subjected to clonogenic assays. Data are representative of three individual experiments. Scale bar, 1 cm. (e) WiDr and HCT116 cells stably transduced with shControl or shINPP4B1 were transduced with a shRNA-resistant mutant form of INPP4B (INPP4B-mut). Forty-eight hours later, whole-cell lysates were subjected to western blotting analysis of INPP4B and GAPDH (as a loading control). Data are representative of three individual experiments. (f) WiDr and HCT116 cells stably transduced with shControl or shINPP4B1 were transduced with a shRNA-resistant mutant form of INPP4B (INPP4B-mut). Forty-eight hours later, cells were subjected to BrdU incorporation assay. Data are represented as mean±s.e.m. of three individual experiments. *P<0.05, Student's t-test. (g) Whole-cell lysates from SW620 and HT-29 colon cancer cells and MDA-MB-231 breast cancer cells stably transduced with the pCDH vector alone or INPP4B cDNA cloned into the pCDH vector (pCDH-INPP4B) were subjected to western blotting analysis of INPP4B, phosphorylated Akt (pSer473-Akt), Akt and GAPDH (as a loading control). Data are representative of three individual experiments. (h) SW620, HT-29 and MDA-MB-231 cells stably transduced with the pCDH vector alone or INPP4B cDNA cloned into the pCDH vector (pCDH-INPP4B) were subjected to clonogenic assays. Data are representative of three individual experiments. Scale bar, 1 cm.
Both Akt and SGK3 contribute to INPP4B-mediated colon cancer cell proliferation
In support of a role of PI3K/Akt activation in INPP4B-mediated colon cancer cell proliferation, INPP4B knockdown reduced phosphorylation of the downstream target glycogen synthase kinase 3β and increased the expression of p27 or p21, two major negative regulators of cell cycle progression that can be inhibited directly or indirectly by PI3K/Akt signaling (Figure 2a and Supplementary Figures S2A and S3A).19 Of note, p21 was not detectable in WiDr and Lim1215 cells with or without knockdown of INPP4B (Supplementary Figure S3A). Nevertheless, introduction of an active form of Akt (myr-Akt) into WiDr and HCT116 cells with INPP4B stably knocked down only partially reversed the inhibitory effect on cell proliferation (Figures 3a and b).
Akt and SGK3 cooperatively regulates colon cancer cell proliferation downstream of INPP4B. (a) WiDr and HCT116 cells stably transduced with the control shRNA (shControl) or INPP4B shRNA (shINPP4B1) were transduced with the vector alone or myr-Akt cDNA. Forty-eight hours later, whole-cell lysates were subjected to western blotting analysis of phosphorylated Akt (pSer473-Akt and pThr308-Akt), Akt and GAPDH (as a loading control). Data are representative of three individual experiments. (b) WiDr and HCT116 cells stably transduced with the shControl or shINPP4B1 were transduced with the vector alone or myr-Akt cDNA. Forty-eight hours later, cells were subjected to bromodeoxyuridine (BrdU) incorporation assays. Data are represented as mean±s.e.m. of three individual experiments. Student's t-test. (c) Whole-cell lysates from WiDr and HCT116 cells stably transduced with shControl or two individual INPP4B shRNAs (shINPP4B1 and shINPP4B2) were subjected to western blotting analysis of phosphorylated SGK3 (pThr320-SGK3), SGK3, phosphorylated SGK1 (pSer422-SGK1), SGK1 and GAPDH (as a loading control). Data are representative of three individual experiments. (d) WiDr and HCT116 cells stably transduced shINPP4B1 were transduced with the vector alone, myr-SGK3 cDNA, myr-Akt cDNA or myr-SGK3 cDNA plus myr-Akt cDNA. Forty-eight hours later, whole-cell lysates were subjected to western blotting analysis of phosphorylated SGK3 (pThr320-SGK3), SGK3, phosphorylated Akt (pSer473-Akt and pThr308-Akt), Akt and GAPDH (as a loading control). Data are representative of three individual experiments. (e) WiDr cells stably transduced with shINPP4B1 were transduced with the vector alone, myr-SGK3 cDNA, myr-Akt cDNA or myr-SGK3 cDNA plus myr-Akt cDNA. Forty-eight hours later, cells were subjected to BrdU incorporation assays. Data are represented as mean±s.e.m. of three individual experiments. *P<0.05, Student's t-test.
We investigated whether SGKs are involved in regulation of colon cancer cell proliferation by INPP4B. INPP4B knockdown caused reduction in phosphorylation (activation) of SGK3 but did not affect phosphorylation of SGK1 (Figure 3c), suggesting that INPP4B preferentially regulates SGK3 activation in colon cancer cells. The functional significance of SGK3 was demonstrated by introduction of an active form of SGK3 (myr-SGK3) into INPP4B knockdown WiDr and HCT116 cells, which, similar to myr-Akt, partially reversed inhibition of cell proliferation by INPP4B knockdown (Figures 3d and e; and Supplementary Figure S3B). When myr-SGK3 and myr-Akt were co-introduced, the inhibitory effect of INPP4B knockdown on cell proliferation was eliminated (Figures 3d and e and Supplementary Figure S3B). Similarly, downregulation of phosphorylated glycogen synthase kinase 3β and upregulation of p27 was abolished (Supplementary Figure S3C).
We also examined the potential role of INPP4B in activation of Akt and SGK3 in colon cancers in vivo. The results showed that, among the 124 colon cancers detailed in Supplementary Table S1, the levels of activated Akt (pSer473-Akt) and SGK3 (pThr320-SGK3) as determined by immunohistochemistry were associated, albeit moderately, with the levels of INPP4B (Supplementary Figure S4).
INPP4B modulates colon cancer growth
To investigate whether INPP4B-mediated activation of PI3K signaling has a role in normal colon epithelial cell transformation, we introduced an INPP4B-expressing construct into FHC cells (Figure 4a). This triggered activation of Akt and SGK3 and caused anchorage-independent growth of the cells (Figures 4a and b). Similarly, it increased the proliferation rate of FHC cells (Figures 4c and d), which could be partially inhibited by co-introduction of a shRNA against Akt or SGK3 (Figures 4e and f and Supplementary Figure S5). When Akt and SGK3 were co-knocked down, the increase in FHC cell proliferation caused by the expression of INPP4B was abolished (Figures 4e and f and Supplementary Figure S5)
INPP4B promotes proliferation and anchorage-independent growth of normal colon epithelial cells and modulates colon cancer xenograft growth. (a) Whole-cell lysates from FHC normal colon epithelial cells stably transduced with the pCDH vector alone or INPP4B cDNA cloned into the pCDH vector (pCDH-INPP4B) were subjected to western blotting analysis of INPP4B, phosphorylated Akt (pSer473-Akt and pThr308-Akt), Akt, phosphorylated SGK3 (pThr320-SGK3), SGK3 and GAPDH (as a loading control). Data are representative of three individual experiments. (b) Representative microphotographs of anchorage-independent growth of FHC cells transduced with the pCDH vector alone or INPP4B cDNA cloned into the pCDH vector (pCDH-INPP4B). Data are representative of three individual experiments. Scale bar, 0.5 mm. (c) FHC cells transduced with the pCDH vector alone or INPP4B cDNA cloned into the pCDH vector (pCDH-INPP4B) were subjected to bromodeoxyuridine (BrdU) incorporation assays. Data are represented as mean±s.e.m. of three individual experiments. *P<0.05, Student's t-test. (d) FHC cells were transduced with the pCDH vector alone or INPP4B cDNA cloned into the pCDH vector (pCDH-INPP4B). Viable cells were counted in an automated cell counter at days 3, 6 and 9 after transduction. Data are represented as mean±s.e.m. of three individual experiments. *P<0.05, Student's t-test. (e) FHC cells transduced with the pCDH vector alone or INPP4B cDNA cloned into the pCDH vector (pCDH-INPP4B) were transduced with shControl, Akt shRNA (shAkt), SGK3 shRNA (shSGK3) or shAKT plus shSGK3. Forty-eight hours later, whole-cell lysates were subjected to western blotting analysis of Akt, SGK3, INPP4B and GAPDH (as a loading control). Data are representative of three individual experiments. (f) FHC cells transduced with the pCDH vector alone or INPP4B cDNA cloned into the pCDH vector (pCDH-INPP4B) were transduced with shControl, shAkt or shSGK3. Forty-eight hours later, cells were subjected to BrdU incorporation assays. Data are represented as mean±s.e.m. of three individual experiments. *P<0.05, Student's t-test. (g) Representative photographs of xenografts of HCT116 cells transduced with shControl or shINPP4B1 in flanks of nu/nu mice (n=8). Mice were euthanized and tumor harvested at 36 days after cell injection. Scale bar, 5 mm. (h) Comparison of growth curves of xenografts of HCT116 cells transduced with shControl or shINPP4B1. Data are represented as mean±s.e.m. of xenografts in 8 mice. *P<0.05; **P<0.01, Student's t-test. (i) Comparison of weight of harvested xenografts of HCT116 cells transduced with shControl or shINPP4B1. Data are represented as mean±s.e.m. of xenografts in eight mice. **P<0.01, Student's t-test. (j) Whole-cell lysates of crude tissues from representative xenografts of HCT116 cells transduced with shControl or shINPP4B1 were subjected to western blotting analysis of INPP4B, phosphorylated Akt (pSer473-Akt), Akt, phosphorylated SGK3 (pThr320-SGK3), SGK3, p27, p21 and GAPDH (as a loading control). Data shown are representative of three individual western blotting analyses of randomly selected tumor samples. (k) HCT116 cells transduced with shINPP4B1 were transduced with the vector alone, myr-Akt cDNA or myr-SGK3 cDNA. Forty-eight hours later, whole-cell lysates were subjected to western blotting analysis of phosphorylated Akt (pSer473-Akt), Akt, phosphorylated SGK3 (pThr320-SGK3), SGK3 and GAPDH (as a loading control). Data shown are representative of three individual experiments. (l) Representative photographs of xenografts of HCT116 cells transduced with shINPP4B1 with or without co-transduction with the vector alone, myr-Akt cDNA or myr-SGK3 cDNA into the flanks of nu/nu mice (n=8). Mice were euthanized and tumor harvested at 36 days after cell injection. Scale bar, 5 mm. (m) Comparison of volume of xenografts of HCT116 cells transduced with shINPP4B1 with or without co-transduction with the vector alone, myr-Akt cDNA or myr-SGK3 cDNA. Data are represented as mean±s.e.m. of xenografts in eight mice. *P<0.05, Student's t-test. (n) Comparison of weight of harvested xenografts of HCT116 cells transduced with shINPP4B1 with or without co-transduction with the vector alone, myr-Akt cDNA or myr-SGK3 cDNA. Data are represented as mean±s.e.m. of xenografts in eight mice. *P<0.05, Student's t-test.
To examine whether the expression of INPP4B affects colon cancer growth in vivo, we transplanted HCT116 cells with or without INPP4B stably knocked down into nu/nu mice. Deficiency in INPP4B retarded tumor growth in vivo (Figures 4g–i), which was associated with reduction in Akt and SGK3 activation and elevation in p27 and p21 expression (Figure 4j). The role of inhibition of Akt and SGK3 in suppression of colon cancer growth was confirmed by transplanting INPP4B knockdown HCT116 cells co-introduced with myr-Akt or myr-SGK3 into nu/nu mice (Figures 4k–n).
INPP4B downregulates PTEN in colon cancer cells
We tested the impact of INPP4B on cellular levels of PI(3,4,5)P3, PI(3,4)P2 and PI(3)P. Strikingly, INPP4B knockdown decreased all these phospholipids in WiDr and HCT116 cells (Figures 5a–c), which could be reversed by introduction of the shRNA-resistant form of INPP4B into the cells (Supplementary Figures S6A–C). Similarly, introduction of exogenous INPP4B into SW620 and HT-29 cells increased PI(3,4,5)P3, PI(3,4)P2 and PI(3)P levels (Supplementary Figures S7A–C).
INPP4B downregulates PTEN in colon cancer cells. (a) The relative abundance of PI(3,4,5)P3 in WiDr and HCT116 cells transduced with the control shRNA (shControl), INPP4B shRNA (shINPP4B1), PTEN shRNA (shPTEN) or shINPP4B1 plus shPTEN was measured by using ELISA in lipid extractions of whole cells. Data are represented as mean±s.e.m. of three individual experiments. **P<0.01, Student's t-test. (b) The relative abundance of PI(3,4)P2 in WiDr and HCT116 cells transduced with shControl, shINPP4B1, shPTEN or shINPP4B1 plus shPTEN was measured by using ELISA in lipid extractions of whole cells. Data are represented as mean±s.e.m. of three individual experiments. *P<0.05, Student's t-test. (c) The relative abundance of PI(3)P in WiDr and HCT116 cells transduced with shControl, shINPP4B1, shPTEN or shINPP4B1 plus shPTEN was measured by using ELISA in lipid extractions of whole cells. Data are represented as mean±s.e.m. of three individual experiments. *P<0.05, Student's t-test. (d) Whole-cell lysates from WiDr and HCT116 cells stably transduced with shControl or two INPP4B shRNAs (shINPP4B1 and shINPP4B2) were subjected to western blotting analysis of PTEN, PIB5PA, SHIP2 and GAPDH (as a loading control). Data are representative of three individual experiments. (e) Whole-cell lysates from WiDr and HCT116 cells stably transduced with shControl or shINPP4B1 were subjected to immunoprecipitation with a rabbit antibody against PTEN or control rabbit IgG. The precipitates were subjected to western blotting analysis of PTEN. Data are representative of three individual experiments. (f) Whole-cell lysates from WiDr and HCT116 cells stably transduced with shControl or shINPP4B1 were subjected to immunoprecipitation with a rabbit antibody against PTEN. The precipitated PTEN was subjected to analysis of PTEN lipid phosphatase activity using a PTEN activity ELISA kit. The relative PTEN lipid phosphatase activity in cells transduced with shControl was arbitrarily designated as 1. Data are represented as mean±s.e.m. of three individual experiments. *P<0.05, Student's t-test. (g) Whole-cell lysates from WiDr and HCT116 cells transduced with shControl, shINPP4B1, shPTEN or shINPP4B1 plus shPTEN were subjected to western blotting analysis of INPP4B, PTEN, hosphorylated Akt (pSer473-Akt), Akt, phosphorylated SGK3 (pThr320-SGK3), SGK3 and GAPDH (as a loading control). Data are representative of three individual experiments. (h) WiDr and HCT116 cells were transduced with shControl, shINPP4B1, shPTEN or shINPP4B1 plus shPTEN. Forty-eight hours later, cells were then subjected to bromodeoxyuridine (BrdU) incorporation assays. Data are represented as mean±s.e.m. of three individual experiments. *P<0.05, Student's t-test.
INPP4B does not hydrolyze PI(3,4,5)P3.9 On the other hand, cellular PI(3,4,5)P3 levels are reduced by PTEN and 5-phosphatases.11, 12, 13 Intriguingly, INPP4B knockdown upregulated PTEN, but did not alter the levels of PIB5PA and SHIP2, in WiDr and HCT116 cells (Figure 5d). Moreover, overexpression of INPP4B downregulated PTEN in SW620 and HCT116 cells (Supplementary Figure S7D). These results suggest that promotion of PI3K signaling by INPP4B is associated with suppression of PTEN in colon cancer cells.
Upregulation of PTEN by INPP4B knockdown in colon cancer cells was associated with an increase in the lipid phosphatase activity of PTEN (Figures 5e and f). Indeed, co-knockdown of PTEN reversed downregulation of PI(3,4,5)P3 and abolished inhibition of Akt and SGK3 activation caused by INPP4B knockdown in WiDr and HCT116 cells (Figures 5a and g). Moreover, it eliminated INPP4B knockdown-induced inhibition of cell proliferation (Figure 5h). Of note, co-knockdown of PTEN did not significantly affect the changes in the levels of PI(3,4)P2 and PI(3)P caused by INPP4B knockdown (Figures 5b and c). Nevertheless, knockdown of PTEN caused increases in the INPP4B expression in WiDr and HCT116 cells (Figure 5g).
The negative regulatory effect of INPP4B on the expression of PTEN was further supported by examination of the relationship between the expression levels of the proteins in colon cancer tissues. The results showed that, among colon cancers that had detectable levels of PTEN expression (76% of the 124 cases; Supplementary Table S1), those with high levels of INPP4B defined using the median of INPP4B levels as the cutoff point displayed reduced levels of PTEN (Supplementary Figure S4).
Downregulation of PTEN by INPP4B is mediated by its protein phosphatase activity
INPP4B knockdown did not affect the levels of the PTEN mRNA (data not shown). Moreover, the half-life of the PTEN protein was prolonged by INPP4B knockdown in colon cancer cells (Figure 6a). These results suggest that INPP4B knockdown-triggered PTEN upregulation is due to a posttranslational increase. In support, the increase in PTEN caused by INPP4B knockdown was associated with increased phosphorylation of the Ser380, Thr382, Thr383 and Ser385 cluster of the protein (Figure 6b), which is known to increase PTEN half-life, in colon cancer cells.20
Downregulation of PTEN by INPP4B is mediated by its protein phosphatase activity. (a) WiDr and HCT116 cells stably transduced with the control shRNA (shControl) or INPP4B shRNA (shINPP4B1) were treated with cycloheximide (CHX) (100 μg/ml) for the indicated periods. Whole-cell lysates were subjected to western blotting analysis of PTEN and GAPDH (as a loading control). Data are representative of three individual experiments. (b) Whole-cell lysates from WiDr and HCT116 cells stably transduced with shControl or two individual INPP4B shRNAs (shINPP4B1 and shINPP4B2) were subjected to western blotting analysis of phosphorylated PTEN (pSer380/Thr382/383 and pSer385), PTEN and GAPDH (as a loading control). Data are representative of three individual experiments. (c) Whole-cell lysates from WiDr and HCT116 cells were subjected to immunoprecipitation with a rabbit antibody against INPP4B or rabbit IgG. Precipitated INPP4B was then subjected to analysis of INPP4B protein Tyrosine phosphatase activity using a Tyrosine phosphatase assay system. The relative INPP4B protein Tyrosine phosphatase activity in cells precipitated with rabbit IgG was arbitrarily designated as 1. Data are represented as mean±s.e.m. of three individual experiments. (d) Whole-cell lysates from WiDr and HCT116 cells were subjected to immunoprecipitation with a rabbit antibody against INPP4B or rabbit IgG. Precipitated INPP4B was then subjected to analysis of INPP4B protein Serine/Threonine phosphatase activity using a Serine/Threonine phosphatase assay system. The relative INPP4B protein Serine/Threonine phosphatase activity in cells precipitated with rabbit IgG was arbitrarily designated as 1. Data are represented as mean±s.e.m. of three individual experiments. (e) Whole-cell lysates from WiDr and HCT116 cells were subjected to immunoprecipitation with a rabbit antibody against INPP4B or rabbit IgG. The resulting precipitates were subjected to western blotting analysis of INPP4B and phosphorylated PTEN (pSer380/Thr382/383). Data are representative of three individual experiments. (f) Whole-cell lysates from WiDr and HCT116 cells were subjected to immunoprecipitation with a rabbit antibody against INPP4B in the absence or presence of NaF (50 mM). The resulting precipitates were then subjected to analysis of INPP4B protein Serine/Threonine phosphatase activity using a Serine/Threonine phosphatase assay system. Data are represented as mean±s.e.m. of three individual experiments. **P<0.01, Student's t-test. (g) Whole-cell lysates from SW620 cells stably transduced with the wild-type INPP4B cDNA (INPP4B-wt) or mutant INPP4B cDNAs (INPP4B-C842S or INPP4B-K846M) cloned into the pCDH vector were subjected to western blotting analysis of INPP4B, PTEN, phosphorylated PTEN (pSer380/Thr382/383), PTEN, phosphorylated Akt (pSer473-Akt), Akt, phosphorylated SGK3 (pThr320-SGK3), SGK3 and GAPDH (as a loading control). Data are representative of three individual experiments. (h) The relative abundance of PI(3,4,5)P3 in SW620 cells transduced with the wild-type INPP4B cDNA (INPP4B-wt) or mutant INPP4B cDNAs (INPP4B-C842S or INPP4B-K846M) cloned into the pCDH vector was measured by using ELISA in lipid extractions of whole cells. Data are represented as mean±s.e.m. of three individual experiments. *P<0.05, Student's t-test. (i) The relative abundance of PI(3,4)P2 in SW620 cells transduced with the wild-type INPP4B cDNA (INPP4B-wt) or mutant INPP4B cDNAs (INPP4B-C842S or INPP4B-K846M) cloned into the pCDH vector was measured by using ELISA in lipid extractions of whole cells. Data are represented as mean±s.e.m. of three individual experiments. *P<0.05, Student's t-test. (j) Whole-cell lysates from HCT116 cells with INPP4B stably knocked down were subjected to GST pull-down using GST-INPP4B, GST-INPP4B-C842S, GST-INPP4B-K846M or GST as bait in the presence of NaF (10 mm). The resulting proteins were then subjected to western blotting analysis of phosphorylated PTEN (pSer380/Thr382/383), GST and PTEN (input control). Data are representative of three individual experiments. (k) Whole-cell lysates from HCT116 cells with INPP4B stably knocked down were incubated with purified GST-INPP4B, GST-INPP4B-C842S, GST-INPP4B-K846M or GST protein for 16 h. Samples were then subjected to western blotting analysis of phosphorylated PTEN (pSer380/Thr382/383), GST and PTEN (input control). Data are representative of three individual experiments.
INPP4B contains a dual specificity phosphatase domain at its catalytic site.21 Indeed, it displayed protein tyrosine (Tyr) and serine/threonine (Ser/Thr) phosphatase activity in colon cancer cells (Figures 6c–e).21 The Ser/Thr phosphatase activity was confirmed by using the Ser/Thr phosphatase inhibitor sodium fluoride (NaF; Figure 6f).22 In support, introduction of a phosphatase dead mutant of INPP4B (INPP4B-C842S), but not expression of the K846M mutant that retained the INPP4B protein phosphatase activity but did not have the lipid phosphatase activity,21 abolished the inhibitory effect of INPP4B on the phosphorylation and expression of PTEN, activation of Akt and SGK3 and upregulation of PI(3,4,5)P3 in SW620 cells (Figures 6g and h). Of note, neither of the mutants affected PI(3,4)P2 levels (Figure 6i).
To confirm that INPP4B dephosphorylates PTEN, we carried out glutathione S-transferase (GST) pull-down assays on whole-cell lysates from HCT116 cells with INPP4B stably knocked down using GST-INPP4B, GST-INPP4B-C842S, GST-INPP4B-K846M or GST as bait. NaF was added into the reaction to prevent dephosphorylation and subsequent degradation of PTEN. As anticipated, PTEN phosphorylated at the Ser380, Thr382 and Thr383 cluster was readily detected by western blotting among proteins pulled down with GST-INPP4B and GST-INPP4B-K846M but not among those by GST or GST-INPP4B-C842S (Figure 6j). However, the addition of GST-INPP4B or GST-INPP4B-K846M but not GST or GST-INPP4B-C842S into whole-cell lysates from INPP4B knockdown HCT116 cells in the absence of NaF caused marked reduction of PTEN phosphorylated at the cluster (Figure 6k).
Ets-1 transcriptionally regulates INPP4B in colon cancer cells
As INPP4B protein expression seems largely correlated with its mRNA expression in colon cancer cells (Figures 1e and f; and Supplementary Figure S1A), it is likely that INPP4B is upregulated by transcriptional mechanisms. We examined the expression of a panel of 84 transcription factors using a qPCR-based array in WiDr cells that expressed relatively high levels of the INPP4B mRNA in comparison with SW620 cells that had relatively low levels (Figure 1f). Among the transcription factors that were differentially expressed (Supplementary Table S4), Ets-1 was expressed 11.6-fold higher in WiDr than SW620 cells. The increased expression of Ets-1 in colon cancer cells with high levels of INPP4B was confirmed in the panel of colon cancer cell lines (Figures 7a and b).22 By in silico analysis, we identified three potential Ets-1-binding sites that were clustered at the −279/−26 region of the INPP4B promoter (Supplementary Figure S8A). Indeed, siRNA knockdown of Ets-1 caused reduction in INPP4B expression in WiDr and HCT116 cells (Figure 7c). In contrast, introduction of an Ets-1-expressing construct resulted in upregulation of INPP4B in SW620 and normal colon epithelial cells (Figure 7d). Moreover, chromatin immunoprecipitation (ChIP) assays showed that Ets-1 was associated with the INPP4B promoter in WiDr and HCT116 cells (Figure 7e). When the Ets-1-binding site-enriched fragment was deleted, the INPP4B promoter activity was significantly reduced even when Ets-1 was overexpressed in SW620 and FHC cells (Figure 7f; and Supplementary Figures S8B and C). The functional significance of regulation of INPP4B by Ets-1 was demonstrated by inhibition of WiDr and HCT116 cell proliferation with siRNA knockdown of Ets-1, which was nevertheless reversed, at least partially, by co-introduction of INPP4B (Supplementary Figure S9).
Ets-1 regulates INPP4B in colon cancer cells. (a) Total RNA from the FHC normal colon epithelial cell line and indicated colon cancer cell lines were subjected to qPCR analysis of Ets-1 mRNA expression. The relative abundance of Ets-1 mRNA in FHC cells was arbitrarily designated as 1. Data are represented as mean±s.e.m. of three individual experiments. (b) Whole-cell lysates from the FHC normal colon epithelial cell line and indicated colon cancer cell lines were subjected to western blotting analysis of Ets-1 and GAPDH (as a loading control). Data are representative of three individual experiments. (c) Whole-cell lysates from WiDr and HCT116 cells transfected with the control siRNA or two individual Ets-1 siRNAs (Ets-1 siRNA1 and Ets-1 siRNA2) were subjected to western blotting analysis of Ets-1, INPP4B and GAPDH (as a loading control). Data are representative of three individual experiments. (d) Whole-cell lysates from SW620 and the FHC cells transfected with the vector alone or Ets-1 cDNA were subjected to western blotting analysis of Ets-1, INPP4B and GAPDH (as a loading control). Data are representative of three individual experiments. (e) Formaldehyde-cross-linked chromatin of WiDr and HCT116 cells were subjected to immunoprecipitation with an antibody against Ets-1. The precipitates were subjected to PCR amplification using primers for the −279/−26 region of the INPP4B promoter. Data are representative of three individual experiments. (f) WiDr and HCT116 cells were transiently transfected with the pGL3-basic-based reporter constructs (pGL3-vector, pGL3-INPP4B wt or pGL3-INPP4B mut as shown in Supplementary Figure S8B) as well as pRL-TK Renilla luciferase control vector. Twenty-four hours later, cells were subjected to the measurement of the luciferase activity. Data are represented as mean±s.e.m. of three individual experiments. **P<0.01, Student's t-test.
Discussion
INPP4B is emerging as a tumor suppressor in a number of cancers, including basal-like breast cancer and those arising from prostate and ovary through inhibition of PI3K/Akt signaling.8, 15 However, we found in this study that INPP4B promoted Akt activation in colon cancer cells. This is perplexing, as INPP4B has been well demonstrated to dephosphorylate PI(3,4)P2, thus terminating PI3K signaling in many types of cells.8, 9 Nevertheless, INPP4B knockdown reduced, whereas its overexpression increased PI(3,4,5)P3 and PI(3,4)P2 levels, suggesting that INPP4B promotes the PI3K/Akt pathway through upregulation of cellular levels of PI(3,4,5)P3 and PI(3,4)P2 in colon cancer cells, in contrast to its negative regulation of the pathway through hydrolyzing PI(3,4)P2 in other types of cells.9, 15 Yet, it remains possible that INPP4B also exerts dephosphorylating effect on PI(3,4)P2 in colon cancer cells, but this is conceivably overridden by the increased biogenesis of PI(3,4)P2 owing to dephosphorylation of elevated PI(3,4,5)P3 by 5-phosphatases such as PIB5PA and SHIP2.23, 24
How does INPP4B upregulate PI(3,4,5)P3 and PI(3,4)P2 in colon cancer cells? Our results revealed that upregulation of PI(3,4,5)P3 by INPP4B was due to suppression of PTEN by the protein Ser/Thr phosphatase activity of INPP4B that dephosphorylated PTEN, which is known to reduce the stability of PTEN (Supplementary Figure S10).20 However, a paradox arising from these results is that native (non-phosphorylated) PTEN has stronger lipid phosphatase activity than its phosphorylated form,20 which would otherwise exert enhanced hydrolyzing effect on PI(3,4,5)P3 leading to its reduction. Nevertheless, our results clearly showed that the lipid phosphatase activity of PTEN was negatively regulated by INPP4B, suggesting that in colon cancer cells the decrease in the activity resulting from INPP4B-mediated downregulation of PTEN is overwhelming.
Similar to PTEN, the catalytic site of INPP4B contains a dual specificity phosphatase domain that is highly conserved across species, suggestive of functional significance.21 Indeed, protein Tyr phosphatase activity of INPP4B has been recently uncovered.21 Our results showed that INPP4B also had protein Ser/Thr phosphatase activity that was responsible for INPP4B-mediated dephosphorylation of PTEN in colon cancer cells. Exogenous INPP4B could pull down and dephosphorylate endogenous PTEN, suggesting that effect of INPP4B on PTEN in colon cancer cells is not due to cell-type-specific characteristics of INPP4B per se. However, whether INPP4B similarly impinges on PTEN in other types of cells such as basal-like breast cancers remains to be defined, but the inhibitory effect of INPP4B on the PI3K/Akt pathway suggests that it is unlikely to regulate PTEN negatively in these types of cells.8, 15 Noticeably, knockdown of PTEN increased the INPP4B expression in colon cancer cells, suggesting that PTEN may also negatively regulate INPP4B through its protein phosphatase activity. The functional significance of this finding remains to be determined. Regardless, our results have clearly demonstrated that negative regulation of PTEN by INPP4B has an important role in promoting PI3K signaling in colon cancer cells.
An important finding of this study was that SGK3 along with Akt contributed to colon cancer cell proliferation driven by INPP4B. Similar to its effect on PI(3,4,5)P3 and PI(3,4)P2, INPP4B also upregulates PI(3)P in colon cancer cells. This is conceivably associated with increased PI(3,4)P2 available for hydrolysis by INPP4B and contributes to enhanced activation of SGK3 by targeting its early endosomes.6, 7 SGK3 is involved in the pathogenesis of various cancers.6, 7 Of particular interest, it has been recently shown to mediate INPP4B-dependent proliferation of breast cancer cells with low Akt and contribute to the growth of mutant BRAF melanomas.16, 25 It seems that the involvement of SGK3 in cancer cell proliferation and survival may be highly cell type and context dependent. Nevertheless, our results indicate that SGK3 contributes to INPP4B-medaited colon cancer cell proliferation irrespective of their mutational statuses of KRAS and PIK3CA.
Genomic alterations such as gene copy number reduction contribute to loss of other inositol polyphosphate phosphatases such as PTEN and PIB5PA in cancer cells.15, 26 Loss of heterozygosity of INPP4B occurs in >50% of human basal-like breast cancers.9 By analogy, the increase in INPP4B in colon cancer cells may directly result from its gene copy number gain. However, analysis of previously published array comparative genomic hybridization data showed that there was no regional copy number gain at chromosome 4q (where INPP4B maps to 4q31) that spanned across the INPP4B gene (142,944,313– 143,768,585).27 We similarly did not observe any INPP4B copy number alteration in colon cancer cells compared with normal colon epithelial cells (not shown). Instead, our results demonstrated that the increase in INPP4B expression in colon cancer cells was due to transcriptional upregulation mediated by the transcription factor Ets-1, which is commonly elevated in colon cancer cells and is involved in the disease development.28, 29 Upregulation of INPP4B by Ets-1 appears to be involved in Ets-1-mediated colon cancer cell proliferation, as inhibition of colon cancer cell proliferation by siRNA knockdown of Ets-1 was reversed, at least partially, by co-introduction of INPP4B. Of interest, Ets-1 is similarly increased and promotes cell proliferation in many other types of cancers, including basal-like breast cancers.30 However, it is apparent that its upregulatory effect on INPP4B is not dominant, in that INPP4B is reduced in basal-like breast cancer cells.8 On the other hand, INPP4B-dependent activation of SGK3 drives tumourigenesis in a subset of breast cancers with low Akt.16 Whether this is associated with regulation of INPP4B by Ets-1 remains to be clarified.
In summary, we have found in this study that INPP4B functions to promote colon cancer cell proliferation through activating Akt and SGK3. The potential oncogenic role of INPP4B in colon cancer is supported by the findings that ectopic expression of INPP4B enhances proliferation and triggers anchorage-independent growth in normal colon epithelial cells. In addition, knockdown of INPP4B retards colon cancer xenograft growth. Despite these findings, activation of Akt and SGK3 appears only moderately associated with the expression of INPP4B in colon cancer in vivo, consistent with the observation that multiple mechanisms are involved in regulation of PI3K signaling.1, 2 Although our results suggest that INPP4B may be targetable in the treatment of colon cancers with high levels of INPP4B expression, given its role as a tumor suppressor in other types of cancers,8, 9, 15 inhibition of INPP4B in vivo needs to be evaluated with great caution. Regardless, our results indicate that the role of INPP4B in the pathogenesis of different types (subtypes) of cancers needs to be defined discretely.
Materials and methods
Lentiviral gene transduction and DNA constructs
The INPP4B, myr-SGK3, myr-Akt, INPP4B-C842S or INPP4B-K846M cDNA was cloned into the lentiviral expression plasmid pCDH-CMV-MCS-EF1-copGFP (Integrated Sciences, Chatswood, NSW, Australia). An INPP4B mutant generated by introducing four silent nucleotide changes in the region complementary to shINPP4B1 was also cloned into the pCDH-CMV-MCS-EF1-copGFP plasmid. Lentiviral packaging was carried out as described previously.31 Transduction efficiency was monitored by detecting green fluorescent protein (GFP) via flow cytometry.
Laser capture microdissection
For microdissection, colon cancer tissues and paired noncancerous colon tissues were obtained from patients undergoing surgical resection. The sample from the tumor tissue was mounted in Tissue-Tek OCT compound (Sakura Finechemicals, Tokyo, Japan) and frozen. Each sample was then cut into 10–20 serial sections with a thickness of 10 μm, and sections were mounted on uncoated glass slides. The parts containing areas of cancer cells were identified using hematoxylin–eosin staining and then microdissected according to the standard laser capture procedure32, 33 using a PixCell II LCM system (Arcturus Engineering, Mountain View, CA, USA/Olympus, Tokyo, Japan). Total RNA was extracted from Laser-captured cell nests by using PicoPure RNA Isolation Kit according to the manufacturer's protocol, including on-column DNase treatment (Qiagen, Chadstone Centre, VIC, Australia).
PI(3,4,5)P3, PI(3,4)P2 and PI(3)P enzyme-linked immunosorbent assay (ELISA)
Cellular PI(3,4,5)P3, PI(3,4)P2 and PI(3)P were respectively extracted and quantitated using PI(3,4,5)P3, PI(3,4)P2 and PI(3)P Mass ELISA kits from Echelon Biosciences (Salt Lake City, UT, USA) according to the manufacturer’s instruction. The results were recorded and analyzed using Synergy 2 multi-detection microplate reader (BioTek, Winooski, VT, USA).
PTEN lipid phosphatase activity assay
Lipid phosphatase activity of PTEN was quantitated using a PTEN activity ELISA kit from Echelon Biosciences as described previously.34 In brief, 30 μl PTEN or the IgG control precipitates were incubated with 30 μl PI(3,4,5)P3 (16 μm) substrate at 37 °C for 2.5 h. The lipid phosphatase activity of PTEN was quantitated by quantitating the amount of PI(4,5)P2. The results were recorded and analyzed using Synergy 2 multi-detection microplate reader (BioTek).
Protein phosphatase activity assay
Protein Serine/Threonine or Tyrosine phosphatase activity of INPP4B was measured by the Serine/Threonine phosphatase assay system or Tyrosine phosphatase assay system from Promega (San Luis Obispo, CA, USA) as described before.35, 36 Briefly, INPP4B or the control goat IgG precipitates from 2 mg whole-cell lysate were incubated with Serine/Threonine or tyrosine phosphopeptides as the substrate. Levels of released free phosphate or PO4 were determined after 10-min incubation at room temperature. INPP4B protein phosphatase activity was calculated by subtracting the nonspecific activity of in IgG precipitates. Naf, an inhibitor of Serine/Threonine phosphatase, was used to validate the assay.
Transcription factor qPCR array
The transcription factor qPCR array was performed by using the Human Transcription Factors RT2 Profiler PCR Array Kit (Qiagen). RNA was purified from WiDr (expressing high levels of INPP4B) and SW620 (expressing low levels of INPP4B) cells by using miRNeasy Mini Kit (Qiagen) with the on-column DNase digestion step according to the manufacturer’s instruction. cDNA was prepared by using the RT2 First Strand Kit and subjected to qPCR analysis in combination with RT2 SYBR Green Mastermixes with an ABI Fast 7900HT sequence detection system.
ChIP assays
ChIP assays were performed using an anti-Ets-1 antibody (Santa Cruz, Murarrie, QLD, Australia) and the EZ-ChIP Kit (Millipore, Bayswater, VIC, Australia) according to the manufacturers' instructions and as described previously.37 In brief, cells were cross-linked with 1% formaldehyde and the bound DNA fragments were subjected to PCR reactions using the following primer pairs: INPP4B, 5′-TGCATTAACTGTGTGGAATGGT-3′ (sense) and 5′-TCCAAATTTTCTTGTCCAAATGTCA-3′ (antisense); and GAPDH (glyceraldehyde 3-phosphate dehydrogenase; control), 5′-TACTAGCGGTTTTACGGGCG-3′(sense) and 5′-TCGAACAGGAGGAGCAGAGAGCGA-3′(antisense).
GST pull down
GST pull down was carried out using a protocol described previously.38 Briefly, BL21 (DE3) cells (Stratagene, Melbourne, VIC, Australia) were transformed with pGEX-3X-INPP4B or with pGEX-3X to express GST-INPP4B or GST alone before preparation of recombinant proteins adsorbed to glutathione sepharose 4B (GE Healthcare, Silverwater, NSW, Australia). Whole-cell lysates from HCT116 cells with INPP4B stably knocked down were incubated with GST or GST-INPP4B in the presence of 10 mm NaF at 4 °C for 2 h before washing the beads four times in lysis buffer. Samples were then eluted with sodium dodecyl sulfate-polyacrylamide gel electrophoresis sample buffer and analyzed by western blotting.
Statistical analysis and data presentation
The Power and Sample Size Program was used to determine the sample size for in vitro experiments. For animal study, Animal Experimentation Ethics Committee Animal Experimentation Sample Size Calculator was used to estimate the sample size, which represents an optimum number to attain statistical significance of P<0.05 with a 90% probability. All data are expressed as mean±s.e.m. of three independent experiments. The two-tailed Student’s t-test was used to analyze the differences for statistical significance between two selected groups with the assumption of normal distribution of data and equal sample variance. Kruskal–Wallis test was carried out for comparing INPP4B expression in colon cancers and paired adjacent noncancerous colon tissues. Correlation between INPP4B and phosphorylated Akt or phosphorylated SGK3 in human colon cancer tissues was examined with Pearson’s correlation. All materials used for, and results generated from, the study were included for statistical analysis, with no randomization or blinding involved. No exclusion of data points was used.
References
Courtney KD, Corcoran RB, Engelman JA . The PI3K pathway as drug target in human cancer. J Clin Oncol 2010; 28: 1075–1083.
Ferte C, Andre F, Soria JC . Molecular circuits of solid tumors: prognostic and predictive tools for bedside use. Nat Rev Clin Oncol 2010; 7: 367–380.
Colakoglu T, Yildirim S, Kayaselcuk F, Nursal TZ, Ezer A, Noyan T et al. Clinicopathological significance of PTEN loss and the phosphoinositide 3-kinase/Akt pathway in sporadic colorectal neoplasms: is PTEN loss predictor of local recurrence? Am J Surg 2008; 195: 719–725.
Majerus PW, Kisseleva MV, Norris FA . The role of phosphatases in inositol signaling reactions. J Biol Chem 1999; 274: 10669–10672.
Park SJ, Itoh T, Takenawa T . Phosphatidylinositol 4-phosphate 5-kinase type I is regulated through phosphorylation response by extracellular stimuli. J Biol Chem 2001; 276: 4781–4787.
Moniz LS, Vanhaesebroeck B . AKT-ing out: SGK kinases come to the fore. Biochem J 2013; 452: e11–e13.
Bruhn MA, Pearson RB, Hannan RD, Sheppard KE . Second AKT: the rise of SGK in cancer signalling. Growth Factors 2010; 28: 394–408.
Fedele CG, Ooms LM, Ho M, Vieusseux J, O'Toole SA, Millar EK et al. Inositol polyphosphate 4-phosphatase II regulates PI3K/Akt signaling and is lost in human basal-like breast cancers. Proc Natl Acad Sci USA 2010; 107: 22231–22236.
Gewinner C, Wang ZC, Richardson A, Teruya-Feldstein J, Etemadmoghadam D, Bowtell D et al. Evidence that inositol polyphosphate 4-phosphatase type II is a tumor suppressor that inhibits PI3K signaling. Cancer Cell 2009; 16: 115–125.
Kisseleva MV, Cao L, Majerus PW . Phosphoinositide-specific inositol polyphosphate 5-phosphatase IV inhibits Akt/protein kinase B phosphorylation and leads to apoptotic cell death. J Biol Chem 2002; 277: 6266–6272.
Carracedo A, Alimonti A, Pandolfi PP . PTEN level in tumor suppression: how much is too little? Cancer Res 2011; 71: 629–633.
Ma K, Cheung SM, Marshall AJ, Duronio V . PI(3,4,5)P3 and PI(3,4)P2 levels correlate with PKB/akt phosphorylation at Thr308 and Ser473, respectively; PI(3,4)P2 levels determine PKB activity. Cell Signal 2008; 20: 684–694.
Ooms LM, Fedele CG, Astle MV, Ivetac I, Cheung V, Pearson RB et al. The inositol polyphosphate 5-phosphatase, PIPP, Is a novel regulator of phosphoinositide 3-kinase-dependent neurite elongation. Mol Biol Cell 2006; 17: 607–622.
Ye Y, Jin L, Wilmott JS, Hu WL, Yosufi B, Thorne RF et al. PI(4,5)P2 5-phosphatase A regulates PI3K/Akt signalling and has a tumour suppressive role in human melanoma. Nat Commun 2013; 4: 1508.
Hodgson MC, Shao LJ, Frolov A, Li R, Peterson LE, Ayala G et al. Decreased expression and androgen regulation of the tumor suppressor gene INPP4B in prostate cancer. Cancer Res 2011; 71: 572–582.
Gasser JA, Inuzuka H, Lau AW, Wei W, Beroukhim R, Toker A . SGK3 mediates INPP4B-dependent PI3K signaling in breast cancer. Mol Cell 2014; 56: 595–607.
Dzneladze I, He R, Woolley JF, Hi Son M, Sharobim MH, Greenberg SA et al. INPP4B overexpression is associated with poor clinical outcome and therapy resistance in acute myeloid leukemia. Leukemia 2015; 29: 1485–1495.
Rijal S, Fleming S, Cummings N, Rynkiewicz NK, Ooms LM, Nguyen NY et al. Inositol polyphosphate 4-phosphatase II (INPP4B) is associated with chemoresistance and poor outcome in AML. Blood 2015; 125: 2815–2824.
Liang J, Slingerland JM . Multiple roles of the PI3K/PKB (Akt) pathway in cell cycle progression. Cell cycle 2003; 2: 339–345.
Hopkins BD, Hodakoski C, Barrows D, Mense SM, Parsons RE . PTEN function: the long and the short of it. Trends Biochem Sci 2014; 39: 183–190.
Lopez SM, Hodgson MC, Packianathan C, Bingol-Ozakpinar O, Uras F, Rosen BP et al. Determinants of the tumor suppressor INPP4B protein and lipid phosphatase activities. Biochem Biophys Res Commun 2013; 440: 277–282.
Jaumot M, Hancock JF . Protein phosphatases 1 and 2 A promote Raf-1 activation by regulating 14-3-3 interactions. Oncogene 2001; 20: 3949–3958.
Franke TF, Kaplan DR, Cantley LC, Toker A . Direct regulation of the Akt proto-oncogene product by phosphatidylinositol-3,4-bisphosphate. Science 1997; 275: 665–668.
Scheid MP, Huber M, Damen JE, Hughes M, Kang V, Neilsen P et al. Phosphatidylinositol (3,4,5)P3 is essential but not sufficient for protein kinase B (PKB) activation; phosphatidylinositol (3,4)P2 is required for PKB phosphorylation at Ser-473: studies using cells from SH2-containing inositol-5-phosphatase knockout mice. J Biol Chem 2002; 277: 9027–9035.
Scortegagna M, Lau E, Zhang T, Feng Y, Sereduk C, Yin H et al. PDK1 and SGK3 contribute to the growth of BRAF mutant melanomas and are potential therapeutic targets. Cancer Res 2015; 75: 1399–1412.
Curtin JA, Busam K, Pinkel D, Bastian BC . Somatic activation of KIT in distinct subtypes of melanoma. J Clin Oncol 2006; 24: 4340–4346.
Guo ST, Jiang CC, Wang GP, Li YP, Wang CY, Guo XY et al. MicroRNA-497 targets insulin-like growth factor 1 receptor and has a tumour suppressive role in human colorectal cancer. Oncogene 2013; 32: 1910–1920.
Ito Y, Takeda T, Okada M, Matsuura N . Expression of ets-1 and ets-2 in colonic neoplasms. Anticancer Res 2002; 22: 1581–1584.
Nakayama T, Ito M, Ohtsuru A, Naito S, Sekine I . Expression of the ets-1 proto-oncogene in human colorectal carcinoma. Mod Pathol 2001; 14: 415–422.
Switzer CH, Cheng RY, Ridnour LA, Glynn SA, Ambs S, Wink DA . Ets-1 is a transcriptional mediator of oncogenic nitric oxide signaling in estrogen receptor-negative breast cancer. Breast Cancer Res 2012; 14: R125.
Dull T, Zufferey R, Kelly M, Mandel RJ, Nguyen M, Trono D et al. A third-generation lentivirus vector with a conditional packaging system. J Virol 1998; 72: 8463–8471.
Bonner RF, Emmert-Buck M, Cole K, Pohida T, Chuaqui R, Goldstein S et al. Laser capture microdissection: molecular analysis of tissue. Science 1997; 278: 1481–1483.
Emmert-Buck MR, Bonner RF, Smith PD, Chuaqui RF, Zhuang Z, Goldstein SR et al. Laser capture microdissection. Science 1996; 274: 998–1001.
Papakonstanti EA, Zwaenepoel O, Bilancio A, Burns E, Nock GE, Houseman B et al. Distinct roles of class IA PI3K isoforms in primary and immortalised macrophages. J Cell Sci 2008; 121: 4124–4133.
Satoh-Nagasawa N, Nagasawa N, Malcomber S, Sakai H, Jackson D . A trehalose metabolic enzyme controls inflorescence architecture in maize. Nature 2006; 441: 227–230.
Nystrom A, Shaik ZP, Gullberg D, Krieg T, Eckes B, Zent R et al. Role of tyrosine phosphatase SHP-1 in the mechanism of endorepellin angiostatic activity. Blood 2009; 114: 4897–4906.
Jin L, Hu WL, Jiang CC, Wang JX, Han CC, Chu P et al. MicroRNA-149*, a p53-responsive microRNA, functions as an oncogenic regulator in human melanoma. Proc Natl Acad Sci USA 2011; 108: 15840–15845.
Struglics A, Fredlund KM, Konstantinov YM, Allen JF, MŁller IM . Protein phosphorylation/dephosphorylation in the inner membrane of potato tuber mitochondria. FEBS Lett 2000; 475: 213–217.
Acknowledgements
This work was supported by the National Health and Medical Research Council (NHMRC) (APP1026458), the Cancer Council NSW (RG13-04 and RG 13-15), Australia, the Shanxi Scholarship Council (2013-133), Shanxi Natural Science Foundation (2013011043-3), China and National Natural Science Foundation of China (81472837). We thank Professor James R Woodgett (Samuel Lunenfeld Research Institute, Canada) for providing the Flag-myristoylated SGK3-CMV construct.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Oncogene website
Supplementary information
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/4.0/
About this article
Cite this article
Guo, S., Chi, M., Yang, R. et al. INPP4B is an oncogenic regulator in human colon cancer. Oncogene 35, 3049–3061 (2016). https://doi.org/10.1038/onc.2015.361
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/onc.2015.361
This article is cited by
-
Tumor innervation is triggered by endoplasmic reticulum stress
Oncogene (2022)
-
Expression and functional characterization of INPP4B in gallbladder cancer patients and gallbladder cancer cells
BMC Cancer (2021)
-
Functional role of SGK3 in PI3K/Pten driven liver tumor development
BMC Cancer (2019)
-
The diacylglycerol kinase α (DGKα)/Akt/NF-κB feedforward loop promotes esophageal squamous cell carcinoma (ESCC) progression via FAK-dependent and FAK-independent manner
Oncogene (2019)
-
Inositol polyphosphate 4-phosphatase type II regulation of androgen receptor activity
Oncogene (2019)