Abstract
Lung cancer is the leading cause of cancer-related death in the United States, and metastatic behavior is largely responsible for this mortality. Mutations in multiple ‘driver’ oncogenes and tumor suppressors are known to contribute to the lung tumorigenesis and in some cases represent therapeutic targets. Leucine Zipper Transcription Factor-like 1 (LZTFL1) is located in the chromosome region 3p21.3 where allelic loss and genetic alterations occur early and frequently in lung cancers. Previously, we found that LZTFL1 is downregulated in epithelial tumors, including lung cancer, and functions as a tumor suppressor in gastric cancers. However, the functional role of LZTFL1 in lung oncogenesis is undefined. We show here that downregulation of LZTFL1 expression in non-small cell lung cancer is associated with recurrence and poor survival, whereas re-expression of LZTFL1 in lung tumor cells inhibited extravasation/colonization of circulating tumor cells to the lung and inhibited tumor growth in vivo. Mechanistically, we found that LZTFL1 is expressed in ciliated human bronchial epithelial cells (HBECs) and its expression correlates with HBEC differentiation. LZTFL1 inhibits transforming growth factor β-activated mitogen-activated protein kinase and hedgehog signaling. Alteration of intracellular levels of LZTFL1 resulted in changes of expression of genes associated with epithelial-to-mesenchymal transition (EMT). We conclude that LZTFL1 inhibits lung tumorigenesis, possibly by maintaining epithelial cell differentiation and/or inhibition of signalings that lead to EMT and suggest that reactivation of LZTFL1 expression in tumor cells may be a novel lung cancer therapeutic approach.
Similar content being viewed by others
Introduction
Lung cancer remains the leading cause of cancer-related death and accounts for 29% of all cancer deaths in the United States despite advances in drug development and surgical procedures. The overall 5-year survival rate of lung cancer is approximately 15%.1, 2 Even for those fortunate to have an early stage diagnosis, 40% will have recurrence after resection and eventually die owing to metastasis.3 The primary cause of lung cancer is smoking that causes somatic mutations of oncogenes and/or tumor suppressors. Research into the underlying mechanisms has identified oncogenic ‘driver’ mutations in genes such as KRAS, EGFR, p53 and others that lead to lung cancer development. This research coupled with genome-wide mutation and epigenetic studies of lung cancer also indicates the existence of additional molecular oncogenic events, including those representing heterogeneity of lung cancer pathogenesis even in tumors with similar key driver mutations. Understanding these additional contributing events hopefully will provide new lung cancer therapeutic opportunities.
Leucine Zipper Transcription Factor-like 1 (LZTFL1) is a gene located in the chromosome region 3p21.3 where allelic loss and genetic alterations occur early and frequently in lung cancers.4, 5 Although LZTFL1 was originally suggested to be a transcription factor based on its sequence similarity to the bHLH family of transcription factor,4 it is a cytoplasmic protein that was shown to interact with other cytosolic proteins regulating ciliary trafficking and controlling β-catenin nuclear signaling.6, 7, 8, 9 Previously, we found that LZTFL1 is expressed in differentiated epithelial cells of a variety of normal tissues, including the lung, and is downregulated in corresponding tumors.6, 7 We showed that LZTFL1 could function as a tumor suppressor in gastric cancers6 and suppresses gastric cancer cell migration and invasion through regulating nuclear translocation of β-catenin, indicating a WNT pathway connection.7 In lung cancer, WNT/β-catenin signaling has been shown to cooperate with the KRAS pathway, resulting in accelerated and more aggressive tumor phenotype in mouse models of lung cancer.8 LZTFL1 inhibits ciliary entry of BBSome that is involved in ciliary trafficking of SMO, thus a negative regulator of hedgehog (Hh) pathway signaling.9 Aberrant activation of Hh signaling pathway has been implicated in the development of a variety of tumors, including lung cancer.10, 11 Hh signaling has been shown to have a cell-autonomous role in initiation and maintenance of small-cell lung cancer.11 Both WNT/β-catenin and Hg signaling pathways are known to activate epithelial-to-mesenchymal transition (EMT),12, 13 a process in which epithelial cells acquire a mesenchymal (fibroblast-like) cell phenotype to become more invasive and metastatic. These data suggest us to test whether LZTFL1 may have a role in the development of EMT and the metastatic phenotype of lung cancer.
In the current study, we show that LZTFL1 is expressed in ciliated human bronchial epithelial cells (HBECs) and upregulation of LZTFL1 in HBECs correlates with epithelial cell differentiation. By contrast, we find that LZTFL1 is downregulated in many lung adenocarcinoma and squamous cell carcinomas, and such loss of LZTFL1 expression is correlated with inferior recurrence and survival rates. Functionally, re-expression of LZTFL1 in lung cancer cells inhibits colonization of circulating tumor cells to the lung in an ex vivo metastasis assay and tumorigenesis in vivo. Mechanistically, we show that re-expression of LZTFL1 downregulated the expression of matrix metalloproteinase 10 (MMP10) and sonic hedgehog (SHH). Knockdown of LZTFL1 in LZTFL1-expressing lung cancer cells resulted in downregulation of epithelial cell marker E-cadherin (CDH1) and upregulation of mesenchymal cell marker, including transcription factor ZEB1. Furthermore, we show that LZTFL1 can interact with BRAF and inhibit the mitogen-activated protein kinase (MAPK) signaling in response to transforming growth factor β (TGFβ). Taken together, our results suggest that LZTFL1 inhibits lung tumorigenesis by maintaining cell differentiation and inhibition of signaling pathways that lead to EMT.
Results
LZTFL1 is downregulated in non-small cell lung carcinoma (NSCLC) and its expression level is associated significantly with non-recurrent events and better survival rate
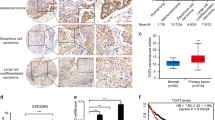
Previously, we found that LZTFL1 is expressed in normal lung epithelium and downregulated in lung cancer on a tissue array by immunohistochemistry (IHC).6 To validate our observation in a larger cohort of patient samples, we performed IHC staining of LZTFL1 in 46 paired normal lung and NSCLC tumor samples obtained from the MD Anderson Cancer Center Thoracic Tissue Bank.14 A strong positive staining of LZTFL1 was observed in the cytoplasm of normal bronchial epithelial cells (Figure 1a). A varying degree of downregulation of cytoplasmic LZTFL1 and alteration in cellular localization were observed in lung adenocarcinoma and squamous cell carcinoma. Cytoplasmic expression of LZTFL1 in adenocarcinoma exhibited a significant inverse correlation with the disease recurrence (Figure 1b). We further examined the association of survival rate with the LZTFL1 expression level in a genome-level transcriptomic meta-database using the online survival analysis software kmplot (www.kmplot.com/lung). This meta-database consists of 10 independent data sets of 1715 samples.15 Higher LZTFL1 level is associated significantly with a higher survival rate in all lung adenocarcinomas (Figure 1c). This survival benefit was also found in never smoking lung adenocarcinoma patients (Figure 1d), whereas no survival benefit was found in smoking lung adenocarcinoma patients (Supplementary Figure S1A). We also analyzed The Cancer Genome Atlas (TCGA) level 3 lung adenocarcinoma and squamous cell carcinoma data sets using X-tile program,16 which allowed us to identify the population subsets based on LZTFL1 expression level along with an associated Kaplan–Meier curve. A significantly higher survival rate associated with higher LZTFL1 expression was also found in adenocarcinoma patients (Figure 1e). No statistically significant survival benefit was observed for squamous cell carcinomas with either kmplot (Supplementary Figure S1B) or X-tile (Supplementary Figure S1C). We also analyzed the copy number variation (CNV) of LZTFL1 in TCGA database. For adenocarcinoma, a higher survival rate is associated with higher CNV number although it did not reach statistically significance (hazard ratio=0.658 (0.420–1.032), logrank P=0.0663; Figure 1f).
LZTFL1 is expressed in normal lung epithelium and downregulated in NSCLCs. (a) LZTFL1 expression in human normal lung epithelium and lung cancers. Clinically annotated, paired normal lung and NSCLC tumor specimens were used to stain LZTFL1 by IHC. Six representative pairs are shown. LZTFL1 is expressed specifically in the cytoplasm of differentiated airway epithelial cells but not in basal cells (arrows). LZTFL1 is downregulated in adeno and squamous cell carcinoma. Interestingly, a membranous and nuclear staining of LZTFL1 can be seen in some lung cancers (arrow head). Scale bar, 40 μM. (b) Cytoplasmic staining of LZTFL1 in adenocarcinoma from non-recurrent (n=13) and recurrent (n=8) patients were scored and subjected to statistical analysis (Pearson r= −0.788, *P<0.01). (c, d) Kaplan–Meier survival plot of lung adenocarcinoma (c, n=719) and never-smoking lung adenocarcinoma patients (d, n=143) using mRNAs of LZTFL1 from an aggregate mega-database and software kmplot. (e, f) Kaplan–Meier survival plot of lung adenocarcinoma patients using LZTFL1 mRNA (e, n=482) and CNV segment mean (f, n=476) from TCGA level 3 data.
LZTFL1 inhibits anchorage-independent lung tumor cell growth
We screened the expression of LZTFL1 in three NSCLC cell lines, and consistent with LZTFL1 being expressed in differentiated lung epithelium, we found that LZTFL1 expression is relatively high in H2347 that is isolated from a well-differentiated primary tumor and low (no expression) in those from metastatic sites (HCC95 and H460) (Figure 2a). H2347 cells are differentiated with expression of the general epithelial cell marker CDH1 and differentiated tumor cell marker KRT1817 (Figure 2a). HCC95 cells are partially differentiated cells that retain the expression of CDH1 but have lost KRT18 expression and also express basal cell marker p63 that is considered to be the stem/progenitor cells of the pulmonary epithelium.18, 19 H460 cells are the least differentiated. To investigate whether LZTFL1 has an active role in the development of NSCLC, we generated several stable cell lines that overexpress either control green fluorescent protein (GFP) or LZTFL1 with bicistronic GFP (LZTFL1-ires-GFP, abbreviated as LZTFL1-GFP hereafter for simplicity) in HCC95 and H460 cells using a lentiviral transduction system. The expression levels of ectopic LZTFL1 in HCC95 and H460 cells are similar to that of endogenous LZTFL1 in normal HBECs (Supplementary Figure S2A). Although overexpression of LZTFL1 did not inhibit tumor cell proliferation in monolayer (data not shown), it partially suppressed the anchorage-independent growth of tumor cells in soft-agar assays (Figure 2b) and inhibited the migratory ability of the cells (Figure 2c).
LZTFL1 inhibits lung tumorigenesis. (a) Western blotting of LZTFL1 and epithelial markers (CDH1 and KRT18) in three lung cancer cell lines. LZTFL1 is expressed in H2347 cells and low (no) in HCC95 and H460 cells. (b) Colony-formation assays with GFP and LZTFL1-GFP expressing H460 and HCC95 cells grown in soft agar. Overexpression of LZTFL1 significantly inhibited colony-formation ability of the tumor cells (n=6±s.e.m.). (c) Relative migration ability of HCC95 and H460 cells transduced with either GFP or LZTFL1-GFP (n=4±s.e.m.). (d) H460-GFP or H460-LZTFL1-GFP cells were IV injected into NOD/SCID mice. Tumors were harvested 6 weeks later. The number of tumor nodules formed in the lung of mice injected with H460-LZTFL1-GFP cells were significantly lower than that of mice injected with control vector transfected cells (H460-GFP) (n=5±s.e.m.). (e) H&E staining of two representative lung sections from each experimental group in (d). (f) Control GFP or LZTFL1-GFP-expressing HCC95 cells were IV injected into a FVB mouse through the tail vein. Lungs were harvested within 15 min of tumor cell injection, sliced and cultured ex vivo. Representative serial images of fluorescent-labeled HCC95 cells in lung sections in the culture dish were taken at the indicated time points with Zeiss fluorescent microscope. (g) Fluorescent colonies were counted and averaged from 10 randomly chosen lung slices/experiment from three independent experiments (n=3±s.e.m.). *P<0.01.
LZTFL1 inhibits lung tumorigenesis in vivo
To test whether LZTFL1 inhibits lung tumorigenesis in vivo, we intravenously (IV) injected H460-GFP or H460-LZTFL1-GFP cells into NOD/SCID mice through the tail vein and followed the tumor progression through bioluminescent imaging using H460 cells expressing a Luciferase reporter (Supplementary Figure S2B). The number of tumor nodules formed in the lungs of mice injected with LZTFL1-GFP-expressing cells was significantly lower than that of mice injected with control vector GFP-expressing cells (Figure 2d). In addition, histological examination showed frequent large dense tumor nodules with severely damaged alveoli in mice injected with GFP cells, while infrequent tumor nodules and tumor cells were seen in mice injected with LZTFL1-GFP cells (Figure 2e).
LZTLF1 inhibits extravasation/colonization of lung tumor cells
The IV injection model contains several steps of metastasis, including tumor cell dissemination through the circulation, extravasation and colonization to the distal tissue/organs. To identify the step(s) that LZTFL1 may inhibit lung tumor metastasis, we tested the effect of LZTFL1 on metastasis of circulating tumor cells in an ex vivo pulmonary metastasis assay.20 HCC95 cells expressing either GFP or LZTFL1-GFP were IV injected into a mouse via tail vein. Metastatic progression from a single cell to a colony in the lung was monitored in real time by GFP-fluorescence in an ex vivo lung culture (Figure 2f). Although similar number of GFP-positive cells were observed between lung tissues injected with control vector GFP-expressing or LZTFL1-GFP-expressing HCC95 cells at 6 h postinjection, significantly lower numbers of GFP-positive cell areas were observed in lung sections injected with LZTFL1-GFP cells 24 and 48 hr postinoculation compared with those injected with GFP-expressing cells (Figure 2g). Similar results were observed for lung tissues injected with H460-GFP and H460-LZTFL1-GFP cells (Supplementary Figure S2C). These data indicate that overexpression of LZTFL1 in tumor cells inhibits the ability of tumor cell to extravasate/colonize the lung in this preclinical model.
LZTFL1 is expressed in ciliated bronchial epithelial cells and its expression is associated with epithelial cell differentiation
As lung epithelium contains multiple cell types, including ciliated (differentiated), undifferentiated columnar, secretory (Clara) and basal cells,21 we co-stained human lungs with LZTFL1 and other cell markers to identify the cell types that express LZTFL1. We observed a graded LZTFL1 staining: low (no) staining in basal (stem/progenitor epithelial cells) cells, no expression in goblet cells, and highest in ciliated (highly differentiated) epithelial cells marked by β-tubulin-IV expression (Figure 3a). To further test whether LZTFL1 expression is associated with epithelial cell differentiation, we measured the transcript level of LZTFL1 in primary HBECs grown in an air–liquid interface (ALI) culture22 in differentiation medium. Little or no LZTFL1 is expressed in the initial 3–7 days after seeding while most of the cells are still in an undifferentiated state. LZTFL1 transcription was upregulated when differentiated ciliated cells were emerging as marked by upregulation of FOXJ123 and reached maximum after ALI day 25 when fully differentiated and beating ciliated cells were present (Figure 3b, Supplementary Figure S3). We were able to partially ‘knock down’ LZTFL1 in differentiating HBECs by infecting HBECs with lentiviruses containing LZTFL1-specific short hairpin RNA (shRNA; Figure 3c). Viral treatment of HBECs resulted in significantly less amount of ciliated HBECs in either control or LZTFL1 shRNA-infected cells compared with non-infected cells (Figure 3d). Although LZTFL1 shRNA downregulation did not block HBEC differentiation as the level of FOXJ1 in LZTFL1 shRNA-infected cells is similar to that of control vector shRNA-treated cells (Supplementary Figure S4), we did observe shorter cilia and fewer ciliated cells in LZTFL1 knockdown cells compared with control vector shRNA knockdown HBECs (Figure 3d, arrows), suggesting LZTFL1 may function downstream of FOXJ1 and may be involved in cilia organization and assemble.
LZTFL1 is expressed in ciliated bronchial epithelial cells and its expression is correlated with epithelial cell differentiation. (a) IHC of human bronchial tissue stained with LZTFL1 (brown color, arrow)) and β-tubulin-IV (arrow head). (b) Relative level of transcripts of LZTFL1 and ciliated epithelial cell marker FOXJ1 normalized against internal GAPDH in ALI culture at the indicated time points during HBEC differentiation. Representative of at least three experiments is shown. (c) Western blotting of LZTFL1 in HBECs infected with none or lentiviruses expressing vector shRNA, or LZTFL1 shRNA. (d) Hematoxylin and eosin-stained sections of none, control shRNA or LZTFL1 shRNA-infected HBECs on inserts at ALI D25. Cilia are marked by arrows. Scale bar=20 μm.
MMP10 is downregulated in LZTFL1-overexpessing cells
To understand the potential mechanism(s) of metastasis inhibition, we studied the whole-genome expression profiles of HCC95-GFP, HCC95-LZTFL1-GFP, H460-GFP and H460-LZTFL1-GFP cells. Of the genes that are more than twofold upregulated or downregulated, we found 95 genes that are altered in both HCC95-LZTFL1-GFP and H460-LZTFL1-GFP cells compared with their respective control GFP-expressing cells (Figure 4a, Supplementary Table S1). MMP10 is the top one gene that is downregulated in HCC95-LZTFL1-GFP cells. As MMP10 has been shown to be involved in cell migration and extravasation/colonization,24 we focused on MMP10 and confirmed its downregulation in LZTFL1-overexprssng HCC95 and H460 cells and in a metastatic breast tumor cell line MDA-MB-231 (Figure 4b). Because LZTFL1 is a cytoplasmic protein, we speculated that MMP10 may not be a direct transcriptional target of LZTFL1 and LZTFL1 may affect a signaling pathway that leads to MMP10 reduction. We cloned the promoter of MMP10 and stimulated H2347 cells with TGFβ as it was shown to activate MMP10 transcription via the MAPK pathway.25 LZTFL1 inhibited TGFβ-activated extracellular signal–regulated kinase (ERK) phosphorylation (Figure 4c). Probing for possible inhibition point(s) along the MAPK pathway, we found that LZTFL1 interacts with BRAF in a co-immunoprecipitation assay (Figure 4d). LZTFL1 inhibited the transcription of MMP10 at baseline and after TGFβ stimulation as well (Figure 4e). However, it failed to inhibit a constitutively active ERK kinase MEK1(R4 F)-activated MMP10 transcription (Figure 4e), suggesting LZTFL1 epistatically acting upstream of MEK1. Taken together, these data suggest that LZTFL1 may inhibit MMP10 expression by suppression of MAPK pathway, possibly via interacting with BRAF (Supplementary Figure S5).
LZTFL1 inhibits tumor cell EMT. (a) Heat map of genes differentially expressed (greater than or equal to twofold) in LZTFL1-expressing HCC95 and H460 cells compared with GFP-expressing cells. (b) Transcripts of MMP10 in LZTFL1-GFP expressing HCC95, H460 and MDA-MB-231 cells measured by qRT–PCR. Transcripts were expressed relative to that in control GFP-expressing cells. (c) H2347 cells were transduced with lentiviral vectors expressing GFP or LZTFL1 and treated with or without TGFβ. Cells were harvested 30 min later for western blotting (WB) analysis with pERK1/2 and ERK1/2 antibodies. (d) myc-BRAF was transfected without or with Flag(f)-LZTFL1 into 293 T cells. Cell lysates were immunoprecipitated (IP) with anti-flag antibody and western blotted with anti-myc antibody. In all, 10% of input was loaded on the gel. (e) MMP10-luciferase activities from H2347 cells transfected with or without LZTFL1, stimulated with or without TGFβ or co-transfected with or without MEK1(R4F). (f, g) Control siRNA (ctl) or LZTFL1-specific siRNAs (LZTFL1-1 and -2) were transfected into H2347 cells. Cell lysates were harvested 72 h later for WB (f) and qRT–PCR analysis (g). For rescuing experiment in panel (f), lentiviruses expressing either GFP or mouse-Lztfl1 were transduced into LZTFL1-siRNA-transfected cells 24 h after transfection.
We further investigated the effect of LZTFL1 knockdown in LZTFL1-expressing H2347 cells (Figure 4f). We used two independent LZTFL1 small interfering RNAs (siRNAs) and rescuing with siRNA-resistant LZTFL1 to address the potential off-target effect of siRNA. Downregulation of LZTFL1 in H2347 cells resulted in decreased expression of epithelial marker CDH1, which can be reversed by adding back ectopically expressed mouse lztfl1 (Figure 4f). Knockdown of LZTFL1 also upregulated the expression of mesenchymal marker ZEB1 (Figure 4g).
LZTFL1 expression inhibits SHH pathway activity in lung cancer cells
As LZTFL1 was shown to be involved in ciliary trafficking and Hh signaling in normal cells, we investigated whether Hh signaling was altered in LZTFL1 exogenously expressed lung tumor cells. Downregulation of SHH in LZTFL1-expressing cells was confirmed by reverse transcriptase–PCR (RT–PCR; (Figure 5a) and western blotting analysis (Figure 5b). Light2 cells contain stably transfected Gli1-luciferase reporter that can be activated in response to Hh signaling.26 We co-cultured control HCC95-GFP or HCC95-LZTFL1-GFP cells with light2 cells. Gli1-luc was downregulated in HCC95-LZTFL1-GFP/light2 co-culture compared with that in HCC95-GFP/light2 co-culture (Figure 5c), confirming that SHH secreted by HCC95-LZTFL1-GFP cells is indeed decreased. We also examined the SHH expression in HCC95-LZTFL1-GFP tumors grown in ex vivo lung culture. SHH in HCC95-LZTFL1-GFP tumors is significantly downregulated compared with that in HCC95-GFP tumors (Figure 5d).
LZTFL1 inhibits Hh signaling. (a) Relative SHH level in HCC95-GFP and HCC95-LZTFL1-GFP cells normalized against internal GAPDH. (b) Western blotting of cell lysates from HCC95-GFP and HCC95-LZTFL1-GFP cells probed with antibodies against SHH and LZTFL1. GAPDH was used as a loading control. (c) Gli1-luc reporter activities from Hh reporter cell line light2 cultured alone, with HCC95-GFP or with HCC95-LZTFL1-GFP cells (n=3±s.e.m., *P<0.05, LZTFL1 vs GFP). (d) IHC of SHH from sections of HCC95-GFP and HCC95-LZTFL1-GFP tumors grown in ex vivo lung culture.
Discussion
The current studies have shown that resected NSCLCs with low expression of the 3p21.3 gene LZTFL1 have inferior survival and that re-expression of LZTFL1 in lung cancers leads to decreased soft-agar colony formation, migratory ability and in vivo metastases formation. By contrast, continued LZTFL1 expression is associated with normal lung epithelial cell differentiation. Thus 3p21.3 allele loss and loss of LZTFL1 expression are functionally important in the malignant behavior of lung cancer. In a most recent TCGA database search, we found that LZTFL1 is homozygously deleted in 1.1% and mutated in 0.6% of lung cancer patients, similar to that of another tumor-suppressor gene RASSF1 in 3p21.3. Majority of the lung cancer patients have lower copy number of LZTFL1 gene. For adenocarcinoma, the survival rate with high CNV segment mean trends to being higher (Figure 1f). The allele loss coupled with probably epigenetic regulation of the remaining allele, downregulation at transcription and posttranscriptional levels or a combination of these mechanism(s) are likely the cause of loss of expression.
The mechanisms by which LZTFL1 inhibits lung cancer metastasis remain to be determined. Based on our studies, we speculate three possibilities. First, LZTFL1 may suppress EMT via inhibition of the MAPK signaling pathways. Several lines of evidence support this. (i) MMP10, CDH1 and ZEB1 are markers associated with EMT. TGFβ is known to promote EMT and regulates the expression of these EMT markers via the MAPK pathways. (ii) Alteration of LZTFL1 in lung tumor cells changes the expression of MMP10, CDH1 and ZEB1 (Figure 4). (iii) LZTFL1 interacts with BRAF and inhibits TGFβ-activated MAPK pathway (Figure 4). That being said, it is noted that evidences for a direct link between LZTFL1, MAPK/SHH pathway and EMT are lacking. Change of the expression of EMT markers may be due to the differentiation status of the cells or due to other secondary events.
Second, LZTFL1 may promote the maintenance of the differentiated state of lung epithelial cells via suppression of the SHH pathways. The association of LZTFL1 with HBEC differentiation appears important. Although knockdown of LZTFL1 did not alter FOXJ1 expression, it did result in shorter cilia and fewer ciliated cells compared with control shRNA-infected cells (Figure 3d), indicating that LZTFL1 acts downstream of FOXJ1 and has a role in final ciliogenesis. LZTFL1 may also be required for maintaining lung epithelium homeostasis by regulating the SHH pathway. SHH signaling has a critical role in lung branching morphogenesis during embryonic lung development. The activity of SHH is decreased during postnatal development and becomes undetectable in the adult lung. SHH signaling is reactivated during lung injuries, lung cancer and cancer cell EMT.27, 28 SHH secreted by epithelial cells can function as a paracrine factor that stimulates mesenchymal proliferation. We found that ectopic expression of LZTFL1 in lung cancer cells inhibits SHH (Figure 5), suggesting that LZTFL1 may have a role in maintaining the differentiated state of lung epithelial cells via suppressing the SHH signaling pathway. That being said, it is noted that the mechanism by which LZTFL1 inhibits SHH production remains to be determined as active SHH is regulated at the level of transcription, ligand processing, secretion or palmitoylation. As a cytoplasmic protein, LZTFL1 could influence the production of active SHH at any one or all of the levels.
Third, suppression of EMT, maintenance of differentiated state and inhibition of the MAPK and SHH signalings by LZTFL1 may be three independent events, although they may interact with each other. LZTFL1 may achieve its metastasis suppression via one or all three mechanisms.
Proteolytic degradation of extracellular matrix by MMPs is a critical step during cancer invasion and metastasis. MMP10 is involved in pulmonary vascular destabilization and remodeling;24 our data show that re-expression of LZTFL1 in lung cancer cells suppresses MMP10 expression, suggesting that LZTFL1 may inhibit extravasation of tumor cells by regulating the expression of genes involved in vascular remodeling. As circulating tumor cells have to extravasate into the lung parenchyma in order to colonize and form subsequent metastases, and extravasation and colonization occur within 2 days of entering the circulation in the mouse model,29 our results suggest that LZTFL1 may inhibit metastasis at the extravasation or colonization step.
Emerging evidence suggest that lung carcinogenesis follows a stepwise model.30 In this model, a malignant transformation starts with a ‘field of cancerization’ and is driven by collective effect of genetic changes of oncogenes and tumor suppressors. Field of cancerization is formed owing to aberrant repair of epithelium by stem/progenitor cells after injury, such as smoking. Genetic and/or epigenetic alterations prevent normal differentiation of these cells, leading to proliferation and expansion of the field and gradually displacing the normal epithelium. Molecular characterization of ‘field of cancerization’ is thus an important research area and the identification of a field effect could be used for risk assessment, early diagnosis and monitoring prevention therapies of lung cancer. Based on the role of LZTFL1 in maintaining epithelial cell differentiation and inhibition of tumorigenesis, it is tempting to speculate that lack of LZTFL1 in the repaired epithelium after injury may mark the area for ‘field of cancerization’.
In summary, our studies suggest that LZTFL1 functions as a lung tumor/metastasis suppressor by promoting/maintaining epithelial cell differentiation and inhibiting EMT-induced MAPK pathway and/or SHH pathway. Developments of therapies that lead to re-expression of LZTFL1 represent a new approach to targeted therapy for lung cancer.
Materials and methods
Plasmid, cell culture, transfection, siRNA and shRNA
The mammalian expression vector pcDNA-myc-LZTFL1 was described previously.6 Human lung tumor cell lines HCC95, H460 and H2347 were established by Minna/Gazdar Laboratory and were maintained in complete medium (RPMI 1640 supplemented with 10% fetal bovine serum, 2 mmol/l glutamine). Cells were transfected with plasmids indicated for each experiment using Lipofectamine 2000 according to the manufacturer's protocol (Invitrogen, Carlsbad, CA, USA). siRNA duplexes were purchased from Sigma (St Louis, MO, USA) and transfected into H2347 cells using Lipofectamine RNA iMAX Reagent (Invitrogen).
Lentiviral transduction
The cDNA of open reading frame of LZTFL1-ires-GFP was PCR amplified from pIres-LZTFL1-GFP plasmid described previously10 and cloned into pSin-E2F-gene-puro vector.31 pSin-E2F-GFP-puro or pSIn-E2F-LZTFL1-ires-GFP-puro were transfected into 293FT cells (Invitrogen) along with packaging vector pMD2-G and envelope vector pSPAX2. Lentiviruses were harvested 36 h later and used to infect HCC95 or H460 cells. Stable cell lines were obtained after puromycin selection. To construct LZTFL1 shRNA lentiviruses, two shRNA sequences of 21 bases corresponding to two different coding regions of LZTFL1 were subcloned in tandem into pLenti6 vector (Invitrogen).
RNA and cDNA preparation, real-time qRT–PCR and microarray analysis
RNA was extracted from cells grown in monolayer with Trizol Reagent according to manufacturer’s instructions (Life Technologies, Carlsbad, CA, USA). First-strand cDNA was made using Superscript III Reverse Transcriptase (Invitrogen). SYBR-based qRT–PCR was used to examine relative levels of selected mRNAs. All data were normalized to an internal standard (GAPDH (glyceraldehyde 3-phosphate dehydrogenase); ΔCT method). Microarray analyses were performed using Illumina bead array platform (San Diego, CA, USA).
Antibodies, western blotting, co-immunoprecipitation and IHC
Anti-LZTFL1 antibody was made in the laboratory and described previously.6 Other antibodies used in this study were obtained commercially; myc, GAPDH (Santa Cruz, Dallas, TX, USA; sc-32233), Flag (Sigma, F3165), CDH1 (E-cad) (Life Technologies, 13-1700), Shh (2287), pERK1/2 (4370), ERK1/2 (7050) (Cell signaling, Danvers, MA, USA), and β-tubulin-IV (Sigma, T5201). Western blottings and co-immunoprecipitation were carried out according to standard protocols. The immunocomplexes on the western blotting were detected by chemiluminescence and photographic films. For IHC staining of LZTFL1 and β-tubulin-IV, human lung tissue sections were deparaffined, probed first with anti-LZTFL1 antibody, detected with horseradish peroxidase/3,3′ diaminobenzidine (dark brown) and re-probed with anti-β-tubulin-IV, followed by AP/NBT (dark blue). The slides were briefly stained with hematoxylin (light blue) for nucleus and mounted with aquamount.
Tissue microarray preparation and IHC staining
Formalin-fixed and paraffin-embedded tissues from clinically annotated, surgically resected lung cancer specimens were obtained from the Lung Cancer Specialized Program of Research Excellence Tissue Bank at MD Anderson Cancer Center used to construct NSCLC tissue microarray. Tissue sections (5 μm) were stained with anti-LZTFL1 antibody and assigned expression scores—staining intensity (0, no staining; 1, weak; 2, medium; 3 strong) multiplied by the proportion of stained cells (1, <50% stained; 2, >50%stained) as described previously.6 All slides were scored by two independent observers blinded to the pathology, and clinical features and averaged values were used for the final LZTFL1 score.
HBEC ALI culture
Human lung tissues were collected after obtaining informed consent on institutional review board-approved protocols. Passage 1 (P1) cells were seeded into plastic dishes in BEGM Basal Epithelial Growth Medium (Lonza, Walkersville, MD, USA; CC-3170). Confluent cultures were passaged to P2 in dishes. After reaching confluence, 1.25 × 105 cells were seeded at 70–90% confluence onto collagen IV (Sigma-Aldrich, St Louis, MO, USA; C7521)-coated 12-mm Transwell permeable supports (BD, Denver, CO, USA; 353180) in ALI culture (STEMCELL Technologies, Vancouver, BC, Canada; 05001). Medium was changed three times a week. For LZTFL1 knockdown experiment, P2 cells at 80% confluence were infected with LZTFL1 shRNA or control enhanced GFP shRNA lentivirus. Cells were selected with 1 μg/ml puromycin 48 h later. After cells were grown back to confluence, cells were passaged and grown subsequently in ALI culture.
IV injection
Cancer cells were harvested at a concentration of 2 × 107 cells/ml of Dulbecco’s modified Eagle’s medium with 10% fetal bovine serum. Of the suspending cells, 0.2 ml was injected through the tail vein into 6-weeek-old NOD/SCID. Tumor growth was monitored by bioluminescence image. Mice were killed after 6 weeks and examined for the growth of metastatic tumors in the lung.
Migration assay and ex vivo pulmonary metastasis assay
Migration assays were performed using 12-well Transwell (Corning, Keller, TX, USA) with 105 cells in Dulbecco’s modified Eagle’s medium on the upper chamber and Dulbecco’s modified Eagle’s medium/10% fetal bovine serum in the lower chamber. Sixteen hours after seeding, cells in the upper chamber were fixed, stained with 4,6-diamidino-2-phenylindole and photographed. Ex vivo metastasis assay was performed according to a published protocol.20 Briefly, 2 × 105 cells were IV injected into the FVB/N mouse via tail vein. The mice were euthanized by CO2 inhalation within 15 min of tumor cell injection. Mouse lungs were exposed, infused with well-mixed culture medium/agarose solution and then removed and placed in a cold solution of phosphate-buffered saline to solidify the agarose/medium solution. Transverse sections were made from each lobe using a no. 21 scalpel blade, were placed on a single sterile Gelfoam (Pfizer-Pharmacia and Upjohn Co.) section in a 6-cm tissue culture dish with culture medium. Lung sections were incubated at 37 °C in humidified conditions of 5% CO2. Fresh culture medium was replaced and lung tissue sections were turned over with a sterile iris thumb forceps every other day. GFP+ tumor cells were photographed using a Zeiss Stemi SV 11 Stereo-Microscope (Zeiss, Pleasanton, CA, USA) equipped with Kramer Scientific Quad-Fluorescence illumination and counted without prior knowledge of the genotype of the cell. All animal studies were approved by the Animal Care and Use Committee of UT Southwestern.
Animals
Male adult (6–8 weeks old) FVB and NOD/SCID mice were used. All animal usage in this study was approved by the Institutional Animal Care and Use Committee of UT Southwestern Medical Center.
Statistics
Student’s t-test (two-tailed) was used to compare the difference between two groups. P<0.05 was considered statistically significant. Pearson’s correlation test and P-value were performed using GraphPad Prism (GraphPad Software, La Jolla, CA, USA). The overall survival analysis in lung cancer patients was conducted using the Kaplan–Meier Plotter webtool (http://www.kmplot.com/lung) and mRNA data from an aggregate mega-database associated with kmplot. The used probe set for LZTFL1 was 218437_s_at; the patient samples are split into two groups according to median expression of the LZTFL1. The two patient cohorts are compared by a Kaplan–Meier survival plot. Univariate Cox regression was performed to compute the hazard ratio with 95% confidence intervals and logrank P-value. Kaplan–Meier survival analysis associated with LZTFL1 mRNA expression value and copy number variation segment mean from TCGA level 3 data were performed by comparing the survival distribution of two groups with logrank test using the X-tile software (http://medicine.yale.edu/lab/rimm/research/software.aspx).16 We used the X-tile software built-in feature that randomly split the patient cohort into a ‘training set’ and a ‘validation set’. The cut point was defined in the training set to achieve the optimal division of outcomes and then independently validated in the ‘validation set’. By doing so, we only tested the difference in the survival outcomes of the groups determined by the cutoff value in the validation set once. The hazard ratio was calculated by COX proportional hazards model.
References
Siegel R, Ward E, Brawley O, Jemal A . Cancer statistics, 2011: the impact of eliminating socioeconomic and racial disparities on premature cancer deaths. CA Cancer J Clin 2011; 61: 212–236.
Edwards BK, Noone AM, Mariotto AB, Simard EP, Boscoe FP, Henley SJ et al. Annual Report to the Nation on the status of cancer, 1975-2010, featuring prevalence of comorbidity and impact on survival among persons with lung, colorectal, breast, or prostate cancer. Cancer 2014; 120: 1290–1314.
Kenny PM, King MT, Viney RC, Boyer MJ, Pollicino CA, McLean JM et al. Quality of life and survival in the 2 years after surgery for non small-cell lung cancer. J Clin Oncol 2008; 26: 233–241.
Kiss H, Kedra D, Kiss C, Kost-Alimova M, Yang Y, Klein G et al. The LZTFL1 gene is a part of a transcriptional map covering 250 kb within the common eliminated region 1 (C3CER1) in 3p21.3. Genomics 2001; 73: 10–19.
Minna JD, Fong K, Zöchbauer-Müller S, Gazdar AF . Molecular pathogenesis of lung cancer and potential translational applications. Cancer J 2002; 8 (Suppl 1): S41–S46.
Wei Q, Zhou W, Wang W, Gao B, Wang L, Cao J et al. Tumor-suppressive functions of leucine zipper transcription factor-like 1. Cancer Res 2010; 70: 2942–2950.
Wang L, Guo J, Wang Q, Zhou J, Xu C, Teng R et al. LZTFL1 suppresses gastric cancer cell migration and invasion through regulating nuclear translocation of β-catenin. J Cancer Res Clin Oncol 2014; 140: 1997–2008.
Pacheco-Pinedo EC, Durham AC, Stewart KM, Goss AM, Lu MM, Demayo FJ et al. Wnt/β-catenin signaling accelerates mouse lung tumorigenesis by imposing an embryonic distal progenitor phenotype on lung epithelium. J Clin Invest 2011; 121: 1935–1945.
Seo S, Zhang Q, Bugge K, Breslow DK, Searby CC, Nachury MV et al. A novel protein LZTFL1 regulates ciliary trafficking of the BBSome and Smoothened. PLoS Genet 2011; 7: e1002358.
Jia Y, Wang Y, Xie J . The Hedgehog pathway: role in cell differentiation, polarity and proliferation. Arch Toxicol 2015; 89: 179–191.
Park KS, Martelotto LG, Peifer M, Sos ML, Karnezis AN, Mahjoub MR et al. A crucial requirement for Hedgehog signaling in small cell lung cancer. Nat Med 2011; 17: 1504–1508.
Maitah MY, Ali S, Ahmad A, Gadgeel S, Sarkar FH . Up-regulation of sonic hedgehog contributes to TGF-β1-induced epithelial to mesenchymal transition in NSCLC cells. PLoS One 2011; 6: e16068.
Xiao D, He J . Epithelial mesenchymal transition and lung cancer. J Thorac Dis 2010; 2: 154–159.
Nanjundan M, Byers LA, Carey MS, Siwak DR, Raso MG, Diao L et al. Proteomic profiling identifies pathways dysregulated in non-small cell lung cancer and an inverse association of AMPK and adhesion pathways with recurrence. J Thorac Oncol 2010; 5: 1894–1904.
Gyorffy B, Surowiak P, Budczies J, Lanczky A . Online survival analysis software to assess the prognostic value of biomarkers using transcriptomic data in non-small-cell lung cancer. PLoS One 2013; 8: e82241.
Camp RL, Dolled-Filhart M, Rimm DL . X-tile: a new bio-informatics tool for biomarker assessment and outcome-based cut-point optimization. Clin Cancer Res 2004; 10: 7252–7259.
Fortier AM, Asselin E, Cadrin M . Keratin 8 and 18 loss in epithelial cancer cells increases collective cell migration and cisplatin sensitivity through claudin1 up-regulation. J Biol Chem 2013; 288: 11555–11571.
Watanabe H, Ma Q, Peng S, Adelmant G, Swain D, Song W et al. SOX2 and p63 colocalize at genetic loci in squamous cell carcinomas. J Clin Invest 2014; 124: 1636–1645.
Zuo W, Zhang T, Wu DZ, Guan SP, Liew AA, Yamamoto Y et al. p63(+)Krt5(+) distal airway stem cells are essential for lung regeneration. Nature 2015; 517: 616–620.
Mendoza A, Hong SH, Osborne T, Khan MA, Campbell K, Briggs J et al. Modeling metastasis biology and therapy in real time in the mouse lung. J Clin Invest 2010; 120: 2979–2988.
Rock JR, Hogan BL . Epithelial progenitor cells in lung development, maintenance, repair, and disease. Annu Rev Cell Dev Biol 2011; 27: 493–512.
Gao X, Vockley CM, Pauli F, Newberry KM, Xue Y, Randell SH et al. Evidence for multiple roles for grainyheadlike 2 in the establishment and maintenance of human mucociliary airway epithelium. Proc Natl Acad Sci USA 2013; 110: 9356–9361.
Look DC, Walter MJ, Williamson MR, Pang L, You Y, Sreshta JN et al. Effects of paramyxoviral infection on airway epithelial cell Foxj1 expression, ciliogenesis, and mucociliary function. Am J Pathol 2001; 159: 2055–2069.
Huang Y, Song N, Ding Y, Yuan S, Li X, Cai H et al. Pulmonary vascular destabilization in the premetastatic phase facilitates lung metastasis. Cancer Res 2009; 69: 7529–7537.
Uttamsingh S, Bao X, Nguyen KT, Bhanot M, Gong J, Chan JL et al. Synergistic effect between EGF and TGF-beta1 in inducing oncogenic properties of intestinal epithelial cells. Oncogene 2008; 27: 2626–2634.
Taipale J, Chen JK, Cooper MK, Wang B, Mann RK, Milenkovic L et al. Effects of oncogenic mutations in Smoothened and Patched can be reversed by cyclopamine. Nature 2000; 406: 1005–1009.
Sato M, Shames DS, Hasegawa Y . Emerging evidence of epithelial-to-mesenchymal transition in lung carcinogenesis. Respirology 2012; 17: 1048–1059.
Kugler MC, Joyner AL, Loomis CA, Munger JS . Sonic hedgehog signaling in the lung. From development to disease. Am J Respir Cell Mol Biol 2015; 52: 1–13.
Gupta GP, Nguyen DX, Chiang AC, Bos PD, Kim JY, Nadal C et al. Mediators of vascular remodelling co-opted for sequential steps in lung metastasis. Nature 2007; 446: 765–770.
Gomperts BN, Walser TC, Spira A, Dubinett SM . Enriching the molecular definition of the airway ‘field of cancerization:’ establishing new paradigms for the patient at risk for lung cancer. Cancer Prev Res (Phila) 2013; 6: 4–7.
Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S et al. Induced pluripotent stem cell lines derived from human somatic cells. Science 2007; 318: 1917–1920.
Acknowledgements
This work is supported by NIHRO1 and CPRIT-MIRA RP120717-P1 (to ZPL), National Natural Science Foundation of China No.81101581 (to QW), National Cancer Institute Specialized Program of Research Excellence in Lung Cancer Grant P50CA70907 (to JDM) and Cancer Center Support Grant by the National Cancer Institute of the National Institutes of Health under award number 5P30CA142543.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Oncogene website
Supplementary information
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/4.0/
About this article
Cite this article
Wei, Q., Chen, ZH., Wang, L. et al. LZTFL1 suppresses lung tumorigenesis by maintaining differentiation of lung epithelial cells. Oncogene 35, 2655–2663 (2016). https://doi.org/10.1038/onc.2015.328
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/onc.2015.328
This article is cited by
-
Role of the Neanderthal Genome in Genetic Susceptibility to COVID-19: 3p21.31 Locus in the Spotlight
Biochemical Genetics (2024)
-
LZTFL1 inhibits kidney tumor cell growth by destabilizing AKT through ZNRF1-mediated ubiquitin proteosome pathway
Oncogene (2023)
-
COVID-19 is not a causal risk for miscarriage: evidence from a Mendelian randomization study
Journal of Assisted Reproduction and Genetics (2023)
-
DNA methylation predicts the outcome of COVID-19 patients with acute respiratory distress syndrome
Journal of Translational Medicine (2022)
-
Genome-wide DNA methylome analysis identifies methylation signatures associated with survival and drug resistance of ovarian cancers
Clinical Epigenetics (2021)