Abstract
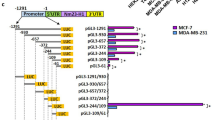
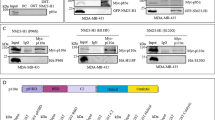
Metastasis is a multistep cell-biological process, which is orchestrated by many factors, including metastasis activators and suppressors. Metastasis Suppressor 1 (MTSS1) was originally identified as a metastasis suppressor protein whose expression is lost in metastatic bladder and prostate carcinomas. However, recent findings indicate that MTSS1 acts as oncogene and pro-migratory factor in melanoma tumors. Here, we identify and characterized a molecular mechanism controlling MTSS1 expression, which impinges on a pro-tumorigenic role of MTSS1 in breast tumors. We found that in normal and in cancer cell lines ΔNp63 is able to drive the expression of MTSS1 by binding to a p63-binding responsive element localized in the MTSS1 locus. We reported that ΔNp63 is able to drive the migration of breast tumor cells by inducing the expression of MTSS1. Notably, in three human breast tumors data sets the MTSS1/p63 co-expression is a negative prognostic factor on patient survival, suggesting that the MTSS1/p63 axis might be functionally important to regulate breast tumor progression.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Yang A, Kaghad M, Wang Y, Gillett E, Fleming MD, Dotsch V et al. p63, a p53 homolog at 3q27-29, encodes multiple products with transactivating, death-inducing, and dominant-negative activities. Mol Cell 1998; 2: 305–316.
Yang A, McKeon F . P63 and P73: P53 mimics, menaces and more. Nat Rev Mol Cell Biol 2000; 1: 199–207.
Levine AJ, Tomasini R, McKeon FD, Mak TW, Melino G . The p53 family: guardians of maternal reproduction. Nat Rev Mol Cell Biol 2011; 12: 259–265.
Giacobbe A, Bongiorno-Borbone L, Bernassola F, Terrinoni A, Markert EK, Levine AJ et al. p63 regulates glutaminase 2 expression. Cell Cycle 2013; 12: 1395–1405.
Flores ER . The roles of p63 in cancer. Cell Cycle 2007; 6: 300–304.
Candi E, Dinsdale D, Rufini A, Salomoni P, Knight RA, Mueller M et al. TAp63 and DeltaNp63 in cancer and epidermal development. Cell Cycle 2007; 6: 274–285.
Gressner O, Schilling T, Lorenz K, Schulze Schleithoff E, Koch A, Schulze-Bergkamen H et al. TAp63alpha induces apoptosis by activating signaling via death receptors and mitochondria. EMBO J 2005; 24: 2458–2471.
Rocco JW, Leong CO, Kuperwasser N, DeYoung MP, Ellisen LW . p63 mediates survival in squamous cell carcinoma by suppression of p73-dependent apoptosis. Cancer Cell 2006; 9: 45–56.
Trink B, Osada M, Ratovitski E, Sidransky D . p63 transcriptional regulation of epithelial integrity and cancer. Cell Cycle 2007; 6: 240–245.
Candi E, Agostini M, Melino G, Bernassola F . How the TP53 family proteins TP63 and TP73 contribute to tumorigenesis: regulators and effectors. Hum Mut 2014; 35: 702–714.
Graziano V, De Laurenzi V . Role of p63 in cancer development. Biochim Biophys Acta 2011; 1816: 57–66.
Melino G . p63 is a suppressor of tumorigenesis and metastasis interacting with mutant p53. Cell Death Differ 2011; 18: 1487–1499.
Su X, Chakravarti D, Cho MS, Liu L, Gi YJ, Lin YL et al. TAp63 suppresses metastasis through coordinate regulation of Dicer and miRNAs. Nature 2010; 467: 986–990.
Ramsey MR, Wilson C, Ory B, Rothenberg SM, Faquin W, Mills AA et al. FGFR2 signaling underlies p63 oncogenic function in squamous cell carcinoma. J Clin Invest 2013; 123: 3525–3538.
Missero C, Antonini D . Crosstalk among p53 family members in cutaneous carcinoma. Exp Dermatol 2014; 23: 143–146.
Wang X, Mori I, Tang W, Nakamura M, Nakamura Y, Sato M et al. p63 expression in normal, hyperplastic and malignant breast tissues. Breast Cancer 2002; 9: 216–219.
Barbareschi M, Pecciarini L, Cangi MG, Macri E, Rizzo A, Viale G et al. p63, a p53 homologue, is a selective nuclear marker of myoepithelial cells of the human breast. Am J Surg Pathol 2001; 25: 1054–1060.
Memmi EM, Sanarico AG, Giacobbe A, Peschiaroli A, Frezza V, Cicalese A et al. p63 sustains self-renewal of mammary cancer stem cells through regulation of Sonic Hedgehog signaling. Proc Nat Acad Sci USA 2015; 112: 3499–3504.
Du Z, Li J, Wang L, Bian C, Wang Q, Liao L et al. Overexpression of DeltaNp63alpha induces a stem cell phenotype in MCF7 breast carcinoma cell line through the Notch pathway. Cancer Sci 2010; 101: 2417–2424.
Chakrabarti R, Wei Y, Hwang J, Hang X, Andres Blanco M, Choudhury A et al. DeltaNp63 promotes stem cell activity in mammary gland development and basal-like breast cancer by enhancing Fzd7 expression and Wnt signalling. Nat Cell Biol 2014; 16: 1004–1015.
Cheung KJ, Gabrielson E, Werb Z, Ewald AJ . Collective invasion in breast cancer requires a conserved basal epithelial program. Cell 2013; 155: 1639–1651.
Machesky LM, Johnston SA . MIM: a multifunctional scaffold protein. J Mol Med 2007; 85: 569–576.
Truong AB, Kretz M, Ridky TW, Kimmel R, Khavari PA . p63 regulates proliferation and differentiation of developmentally mature keratinocytes. Genes Dev 2006; 20: 3185–3197.
Xie F, Ye L, Ta M, Zhang L, Jiang WG . MTSS1: a multifunctional protein and its role in cancer invasion and metastasis. Front Biosci 2011; 3: 621–631.
Parr C, Jiang WG . Metastasis suppressor 1 (MTSS1) demonstrates prognostic value and anti-metastatic properties in breast cancer. Eur J Cancer 2009; 45: 1673–1683.
Lee YG, Macoska JA, Korenchuk S, Pienta KJ . MIM, a potential metastasis suppressor gene in bladder cancer. Neoplasia 2002; 4: 291–294.
Fan H, Chen L, Zhang F, Quan Y, Su X, Qiu X et al. MTSS1, a novel target of DNA methyltransferase 3B, functions as a tumor suppressor in hepatocellular carcinoma. Oncogene 2012; 31: 2298–2308.
Kedmi M, Ben-Chetrit N, Korner C, Mancini M, Ben-Moshe NB, Lauriola M et al. EGF induces microRNAs that target suppressors of cell migration: miR-15b targets MTSS1 in breast cancer. Sci Signal 2015; 8: ra29.
Dawson JC, Timpson P, Kalna G, Machesky LM . Mtss1 regulates epidermal growth factor signaling in head and neck squamous carcinoma cells. Oncogene 2012; 31: 1781–1793.
Xie F, Ye L, Chen J, Wu N, Zhang Z, Yang Y et al. The impact of Metastasis Suppressor-1, MTSS1, on oesophageal squamous cell carcinoma and its clinical significance. J Transl Med 2011; 9: 95.
Mertz KD, Pathria G, Wagner C, Saarikangas J, Sboner A, Romanov J et al. MTSS1 is a metastasis driver in a subset of human melanomas. Nat Commun 2014; 5: 3465.
Mattila PK, Pykalainen A, Saarikangas J, Paavilainen VO, Vihinen H, Jokitalo E et al. Missing-in-metastasis and IRSp53 deform PI(4,5)P2-rich membranes by an inverse BAR domain-like mechanism. J Cell Biol 2007; 176: 953–964.
Bompard G, Sharp SJ, Freiss G, Machesky LM . Involvement of Rac in actin cytoskeleton rearrangements induced by MIM-B. J Cell Sci 2005; 118: 5393–5403.
Wang W, Eddy R, Condeelis J . The cofilin pathway in breast cancer invasion and metastasis. Nat Rev Cancer 2007; 7: 429–440.
Bergholz J, Zhang Y, Wu J, Meng L, Walsh EM, Rai A et al. DeltaNp63alpha regulates Erk signaling via MKP3 to inhibit cancer metastasis. Oncogene 2014; 33: 212–224.
Fukushima H, Koga F, Kawakami S, Fujii Y, Yoshida S, Ratovitski E et al. Loss of DeltaNp63alpha promotes invasion of urothelial carcinomas via N-cadherin/Src homology and collagen/extracellular signal-regulated kinase pathway. Cancer Res 2009; 69: 9263–9270.
Tucci P, Agostini M, Grespi F, Markert EK, Terrinoni A, Vousden KH et al. Loss of p63 and its microRNA-205 target results in enhanced cell migration and metastasis in prostate cancer. Proc Natl Acad Sci USA 2012; 109: 15312–15317.
Cho MS, Chan IL, Flores ER . DeltaNp63 transcriptionally regulates brachyury, a gene with diverse roles in limb development, tumorigenesis and metastasis. Cell Cycle 2010; 9: 2434–2441.
Lee KB, Ye S, Park MH, Park BH, Lee JS, Kim SM . p63-Mediated activation of the beta-catenin/c-Myc signaling pathway stimulates esophageal squamous carcinoma cell invasion and metastasis. Cancer Lett 2014; 353: 124–132.
Yang X, Lu H, Yan B, Romano RA, Bian Y, Friedman J et al. DeltaNp63 versatilely regulates a Broad NF-kappaB gene program and promotes squamous epithelial proliferation, migration, and inflammation. Cancer Res 2011; 71: 3688–3700.
Matin RN, Chikh A, Chong SL, Mesher D, Graf M, Sanza P et al. p63 is an alternative p53 repressor in melanoma that confers chemoresistance and a poor prognosis. J Exp Med 2013; 210: 581–603.
Malatesta M, Peschiaroli A, Memmi EM, Zhang J, Antonov A, Green DR et al. The Cul4A-DDB1 E3 ubiquitin ligase complex represses p73 transcriptional activity. Oncogene 2013; 32: 4721–4726.
Velletri T, Romeo F, Tucci P, Peschiaroli A, Annicchiarico-Petruzzelli M, Niklison-Chirou MV et al. GLS2 is transcriptionally regulated by p73 and contributes to neuronal differentiation. Cell Cycle 2013; 12: 3564–3573.
Peschiaroli A, Scialpi F, Bernassola F, El Sherbini el S, Melino G . The E3 ubiquitin ligase WWP1 regulates DeltaNp63-dependent transcription through Lys63 linkages. Biochem Biophys Res Commun 2010; 402: 425–430.
Amelio I, Gostev M, Knight RA, Willis AE, Melino G, Antonov AV . DRUGSURV: a resource for repositioning of approved and experimental drugs in oncology based on patient survival information. Cell Death Dis 2014; 5: e1051.
Antonov AV, Krestyaninova M, Knight RA, Rodchenkov I, Melino G, Barlev NA . PPISURV: a novel bioinformatics tool for uncovering the hidden role of specific genes in cancer survival outcome. Oncogene 2014; 33: 1621–1628.
Celardo I, Grespi F, Antonov A, Bernassola F, Garabadgiu AV, Melino G et al. Caspase-1 is a novel target of p63 in tumor suppression. Cell Death Dis 2013; 4: e645.
Acknowledgements
We thank the laboratory members for helpful discussion. This work has been supported by MFAG AIRC 15523 grant awarded to AP, and by Medical Research Council (UK), Ministry of Health ‘Rc1.1’ and ‘Rc 1.3’, AIRC Grant IG-15653, AIRC 5xmille (#9979), ‘NCDs Fondazione Roma’ grant awarded to GM.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Oncogene website
Supplementary information
Rights and permissions
About this article
Cite this article
Giacobbe, A., Compagnone, M., Bongiorno-Borbone, L. et al. p63 controls cell migration and invasion by transcriptional regulation of MTSS1. Oncogene 35, 1602–1608 (2016). https://doi.org/10.1038/onc.2015.230
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/onc.2015.230
This article is cited by
-
Functional antagonism between ΔNp63α and GCM1 regulates human trophoblast stemness and differentiation
Nature Communications (2022)
-
Supercharging BRD4 with NUT in carcinoma
Oncogene (2021)
-
ARID2 deficiency promotes tumor progression and is associated with higher sensitivity to chemotherapy in lung cancer
Oncogene (2021)
-
New immunological potential markers for triple negative breast cancer: IL18R1, CD53, TRIM, Jaw1, LTB, PTPRCAP
Discover Oncology (2021)
-
Atypical GATA transcription factor TRPS1 represses gene expression by recruiting CHD4/NuRD(MTA2) and suppresses cell migration and invasion by repressing TP63 expression
Oncogenesis (2018)