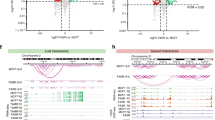
Abstract
Breast cancer resistance to endocrine therapies such as tamoxifen and aromatase inhibitors is a significant clinical problem. Steroid receptor coactivator-1 (SRC-1), a coregulatory protein of the oestrogen receptor (ER), has previously been shown to have a significant role in the progression of breast cancer. The chromatin protein high mobility group box 2 (HMGB2) was identified as an SRC-1 interacting protein in the endocrine-resistant setting. We investigated the expression of HMGB2 in a cohort of 1068 breast cancer patients and found an association with increased disease-free survival time in patients treated with endocrine therapy. However, it was also verified that HMGB2 expression could be switched on in endocrine-resistant tumours from breast cancer patients. To explore the function of this poorly characterized protein, we performed HMGB2 ChIPseq and found distinct binding patterns between the two contexts. In the resistant setting, the HMGB2, SRC-1 and ER complex are enriched at promoter regions of target genes, with bioinformatic analysis indicating a switch in binding partners between the sensitive and resistant phenotypes. Integration of binding and gene expression data reveals a concise set of target genes of this complex including the RNA helicase DDX18. Modulation of DDX18 directly affects growth of tamoxifen-resistant cells, suggesting that it may be a critical downstream effector of the HMGB2:ER complex. This study defines HMGB2 interactions with the ER complex at specific target genes in the tamoxifen-resistant setting.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Davies C, Godwin J, Gray R, Clarke M, Cutter D, Darby S et al. Relevance of breast cancer hormone receptors and other factors to the efficacy of adjuvant tamoxifen: patient-level meta-analysis of randomised trials. Lancet 2011; 378: 771–784.
Onate SA, Tsai SY, Tsai MJ, O'Malley BW . Sequence and characterization of a coactivator for the steroid hormone receptor superfamily. Science 1995; 270: 1354–1357.
Redmond AM, Bane FT, Stafford AT, McIlroy M, Dillon MF, Crotty TB et al. Coassociation of estrogen receptor and p160 proteins predicts resistance to endocrine treatment; SRC-1 is an independent predictor of breast cancer recurrence. Clin Cancer Res 2009; 15: 2098–2106.
Qin L, Liu Z, Chen H, Xu J . The steroid receptor coactivator-1 regulates twist expression and promotes breast cancer metastasis. Cancer Res 2009; 69: 3819–3827.
McBryan J, Theissen SM, Byrne C, Hughes E, Cocchiglia S, Sande S et al. Metastatic progression with resistance to aromatase inhibitors is driven by the steroid receptor coactivator SRC-1. Cancer Res 2012; 72: 548–559.
Walsh CA, Bolger JC, Byrne C, Cocchiglia S, Hao Y, Fagan A et al. Global gene repression by the steroid receptor coactivator SRC-1 promotes oncogenesis. Cancer Res 2014; 74: 2533–2544.
Stros M . HMGB proteins: interactions with DNA and chromatin. Biochim Biophys Acta 2010; 1799: 101–113.
Melvin VS, Harrell C, Adelman JS, Kraus WL, Churchill M, Edwards DP . The role of the C-terminal extension (CTE) of the estrogen receptor alpha and beta DNA binding domain in DNA binding and interaction with HMGB. J Biol Chem 2004; 279: 14763–14771.
Zappavigna V, Falciola L, Helmer-Citterich M, Mavilio F, Bianchi ME . HMG1 interacts with HOX proteins and enhances their DNA binding and transcriptional activation. EMBO J 1996; 15: 4981–4991.
Tremethick DJ, Molloy PL . High mobility group proteins 1 and 2 stimulate transcription in vitro by RNA polymerases II and III. J Biol Chem 1986; 261: 6986–6992.
Laurent B, Randrianarison-Huetz V, Marechal V, Mayeux P, Dusanter-Fourt I, Dumenil D . High-mobility group protein HMGB2 regulates human erythroid differentiation through trans-activation of GFI1B transcription. Blood 2010; 115: 687–695.
Wang W, Jiang H, Zhu H, Zhang H, Gong J, Zhang L et al. Overexpression of high mobility group box 1 and 2 is associated with the progression and angiogenesis of human bladder carcinoma. Oncol Lett 2013; 5: 884–888.
Boonyaratanakornkit V, Melvin V, Prendergast P, Altmann M, Ronfani L, Bianchi ME et al. High-mobility group chromatin proteins 1 and 2 functionally interact with steroid hormone receptors to enhance their DNA binding in vitro and transcriptional activity in mammalian cells. Mol Cell Biol 1998; 18: 4471–4487.
Jolma A, Yan J, Whitington T, Toivonen J, Nitta KR, Rastas P et al. DNA-binding specificities of human transcription factors. Cell 2013; 152: 327–339.
McCartan D, Bolger JC, Fagan A, Byrne C, Hao Y, Qin L et al. Global characterization of the SRC-1 transcriptome identifies ADAM22 as an ER-independent mediator of endocrine-resistant breast cancer. Cancer Res 2012; 72: 220–229.
Grandori C, Mac J, Siebelt F, Ayer DE, Eisenman RN . Myc-Max heterodimers activate a DEAD box gene and interact with multiple E box-related sites in vivo. EMBO J 1996; 15: 4344–4357.
Payne EM, Bolli N, Rhodes J, Abdel-Wahab OI, Levine R, Hedvat CV et al. Ddx18 is essential for cell-cycle progression in zebrafish hematopoietic cells and is mutated in human AML. Blood 2011; 118: 903–915.
Gyorffy B, Lanczky A, Eklund AC, Denkert C, Budczies J, Li Q et al. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res Treat 2010; 123: 725–731.
Musgrove EA, Sutherland RL . Biological determinants of endocrine resistance in breast cancer. Nat Rev Cancer 2009; 9: 631–643.
Badve S, Turbin D, Thorat MA, Morimiya A, Nielsen TO, Perou CM et al. FOXA1 expression in breast cancer–correlation with luminal subtype A and survival. Clin Cancer Res 2007; 13: 4415–4421.
Hisamatsu Y, Tokunaga E, Yamashita N, Akiyoshi S, Okada S, Nakashima Y et al. Impact of FOXA1 expression on the prognosis of patients with hormone receptor-positive breast cancer. Ann Surg Oncol 2012; 19: 1145–1152.
Ross-Innes CS, Stark R, Teschendorff AE, Holmes KA, Ali HR, Dunning MJ et al. Differential oestrogen receptor binding is associated with clinical outcome in breast cancer. Nature 2012; 481: 389–393.
Shiota M, Izumi H, Miyamoto N, Onitsuka T, Kashiwagi E, Kidani A et al. Ets regulates peroxiredoxin1 and 5 expressions through their interaction with the high-mobility group protein B1. Cancer Sci 2008; 99: 1950–1959.
Carroll JS, Meyer CA, Song J, Li W, Geistlinger TR, Eeckhoute J et al. Genome-wide analysis of estrogen receptor binding sites. Nat Genet 2006; 38: 1289–1297.
de Leeuw R, Flach K, Bentin Toaldo C, Alexi X, Canisius S, Neefjes J et al. PKA phosphorylation redirects ERalpha to promoters of a unique gene set to induce tamoxifen resistance. Oncogene 2013; 32: 3543–3551.
Al-azawi D, Ilroy MM, Kelly G, Redmond AM, Bane FT, Cocchiglia S et al. Ets-2 and p160 proteins collaborate to regulate c-Myc in endocrine resistant breast cancer. Oncogene 2008; 27: 3021–3031.
Massarweh S, Osborne CK, Creighton CJ, Qin L, Tsimelzon A, Huang S et al. Tamoxifen resistance in breast tumors is driven by growth factor receptor signaling with repression of classic estrogen receptor genomic function. Cancer Res 2008; 68: 826–833.
Wortham NC, Ahamed E, Nicol SM, Thomas RS, Periyasamy M, Jiang J et al. The DEAD-box protein p72 regulates ERalpha-/oestrogen-dependent transcription and cell growth, and is associated with improved survival in ERalpha-positive breast cancer. Oncogene 2009; 28: 4053–4064.
Limer JL, Parkes AT, Speirs V . Differential response to phytoestrogens in endocrine sensitive and resistant breast cancer cells in vitro. Int J Cancer 2006; 119: 515–521.
Neve RM, Chin K, Fridlyand J, Yeh J, Baehner FL, Fevr T et al. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell 2006; 10: 515–527.
Dillon MF, Stafford AT, Kelly G, Redmond AM, McIlroy M, Crotty TB et al. Cyclooxygenase-2 predicts adverse effects of tamoxifen: a possible mechanism of role for nuclear HER2 in breast cancer patients. Endocr Relat Cancer 2008; 15: 745–753.
Schmidt D, Wilson MD, Spyrou C, Brown GD, Hadfield J, Odom DT . ChIP-seq: using high-throughput sequencing to discover protein-DNA interactions. Methods 2009; 48: 240–248.
Li H, Durbin R . Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009; 25: 1754–1760.
Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol 2008; 9: R137.
Kharchenko PV, Tolstorukov MY, Park PJ . Design and analysis of ChIP-seq experiments for DNA-binding proteins. Nat Biotechnol 2008; 26: 1351–1359.
Li Q, Brown JB, Huang H, Bickel PJ . Measuring reproducibility of high-throughput experiments. Ann Appl Stat 5: 1752–1779.
Liu T, Ortiz JA, Taing L, Meyer CA, Lee B, Zhang Y et al. Cistrome: an integrative platform for transcriptional regulation studies. Genome Biol 2011; 12: R83.
McLean CY, Bristor D, Hiller M, Clarke SL, Schaar BT, Lowe CB et al. GREAT improves functional interpretation of cis-regulatory regions. Nat Biotechnol 2010; 28: 495–501.
Zhang Z, Chang CW, Goh WL, Sung WK, Cheung E . CENTDIST: discovery of co-associated factors by motif distribution. Nucleic Acids Res 2011; 39: W391–W399.
Cairns JM, Dunning MJ, Ritchie ME, Russell R, Lynch AG . BASH: a tool for managing BeadArray spatial artefacts. Bioinformatics 2008; 24: 2921–2922.
Dunning MJ, Smith ML, Ritchie ME, Tavare S . beadarray: R classes and methods for Illumina bead-based data. Bioinformatics 2007; 23: 2183–2184.
Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 2004; 5: R80.
Smyth GK . Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol: 2004; 3; Article 3.
Benjamini Y, Hochberg Y . Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc 1995; 57: 289–300.
Acknowledgements
We would like to acknowledge the support of Irish Research Council for Science Engineering and Technology co-funded by Marie Curie Actions under FP7, Breast Cancer Ireland, Health Research Board, The University of Cambridge, Cancer Research UK, Hutchison Whampoa Limited and European Molecular Biology Organisation. We thank the members of the genomics core facility and Stuart MacArthur, former member of the bioinformatics core facility at Cancer Research UK.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Oncogene website
Supplementary information
Rights and permissions
About this article
Cite this article
Redmond, A., Byrne, C., Bane, F. et al. Genomic interaction between ER and HMGB2 identifies DDX18 as a novel driver of endocrine resistance in breast cancer cells. Oncogene 34, 3871–3880 (2015). https://doi.org/10.1038/onc.2014.323
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/onc.2014.323
This article is cited by
-
Identification of HMGB2 associated with proliferation, invasion and prognosis in lung adenocarcinoma via weighted gene co-expression network analysis
BMC Pulmonary Medicine (2022)
-
Crucial role of high-mobility group box 2 in mouse ovarian follicular development through estrogen receptor beta
Histochemistry and Cell Biology (2022)
-
Identification of differentially expressed proteins and clinicopathological significance of HMGB2 in cervical cancer
Clinical Proteomics (2021)
-
Molecular markers associated with the outcome of tamoxifen treatment in estrogen receptor-positive breast cancer patients: scoping review and in silico analysis
Discover Oncology (2021)
-
HMGB2 is associated with malignancy and regulates Warburg effect by targeting LDHB and FBP1 in breast cancer
Cell Communication and Signaling (2018)