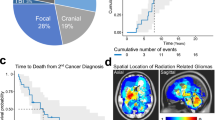
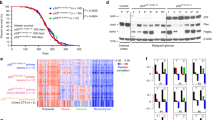
Abstract
Glioblastomas (GBM) are highly radioresistant and lethal brain tumors. Ionizing radiation (IR)-induced DNA double-strand breaks (DSBs) are a risk factor for the development of GBM. In this study, we systematically examined the contribution of IR-induced DSBs to GBM development using transgenic mouse models harboring brain-targeted deletions of key tumor suppressors frequently lost in GBM, namely Ink4a, Ink4b, Arf and/or PTEN. Using low linear energy transfer (LET) X-rays to generate simple breaks or high LET HZE particles (Fe ions) to generate complex breaks, we found that DSBs induce high-grade gliomas in these mice which, otherwise, do not develop gliomas spontaneously. Loss of Ink4a and Arf was sufficient to trigger IR-induced glioma development but additional loss of Ink4b significantly increased tumor incidence. We analyzed IR-induced tumors for copy number alterations to identify oncogenic changes that were generated and selected for as a consequence of stochastic DSB events. We found Met amplification to be the most significant oncogenic event in these radiation-induced gliomas. Importantly, Met activation resulted in the expression of Sox2, a GBM cancer stem cell marker, and was obligatory for tumor formation. In sum, these results indicate that radiation-induced DSBs cooperate with loss of Ink4 and Arf tumor suppressors to generate high-grade gliomas that are commonly driven by Met amplification and activation.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Furnari FB, Fenton T, Bachoo RM, Mukasa A, Stommel JM, Stegh A et al Malignant astrocytic glioma: genetics, biology, and paths to treatment. Genes Dev 2007; 21: 2683–2710.
Westphal M, Lamszus K . The neurobiology of gliomas: from cell biology to the development of therapeutic approaches. Nat Rev Neurosci 2011; 12: 495–508.
Purow B, Schiff D . Advances in the genetics of glioblastoma: are we reaching critical mass? Nat Rev Neurol 2009; 5: 419–426.
Bondy ML, Scheurer ME, Malmer B, Barnholtz-Sloan JS, Davis FG, Il'yasova D et al Brain tumor epidemiology: consensus from the Brain Tumor Epidemiology Consortium. Cancer 2008; 113: 1953–1968.
Ron E . Ionizing radiation and cancer risk: evidence from epidemiology. Pediatr Radiol 2002; 32: 232–237 discussion 42-4.
Neglia JP, Meadows AT, Robison LL, Kim TH, Newton WA, Ruymann FB et al Second neoplasms after acute lymphoblastic leukemia in childhood. New Engl J Med 1991; 325: 1330–1336.
Salvati M, Frati A, Russo N, Caroli E, Polli FM, Minniti G et al Radiation-induced gliomas: report of 10 cases and review of the literature. Surg Neurol 2003; 60: 60–67, discussion 7.
Thierry-Chef I, Simon SL, Land CE, Miller DL . Radiation dose to the brain and subsequent risk of developing brain tumors in pediatric patients undergoing interventional neuroradiology procedures. Radiat Res 2008; 170: 553–565.
Paulino AC, Mai WY, Chintagumpala M, Taher A, Teh BS . Radiation-induced malignant gliomas: is there a role for reirradiation? Int J Radiat Oncol Biol Phys 2008; 71: 1381–1387.
Pearce MS, Salotti JA, Little MP, McHugh K, Lee C, Kim KP et al Radiation exposure from CT scans in childhood and subsequent risk of leukaemia and brain tumours: a retrospective cohort study. Lancet 2012; 380: 499–505.
Durante M, Cucinotta FA . Heavy ion carcinogenesis and human space exploration. Nat Rev Cancer 2008; 8: 465–472.
Okayasu R . Repair of DNA damage induced by accelerated heavy ions—a mini review. Int J Cancer 2012; 130: 991–1000.
Sage E, Harrison L . Clustered DNA lesion repair in eukaryotes: relevance to mutagenesis and cell survival. Mutat Res 2011; 711: 123–133.
Khanna KK, Jackson SP . DNA double-strand breaks: signaling, repair and the cancer connection. Nat Genet 2001; 27: 247–254.
Camacho CV, Mukherjee B, McEllin B, Ding LH, Hu B, Habib AA et al Loss of p15/Ink4b accompanies tumorigenesis triggered by complex DNA double-strand breaks. Carcinogenesis 2010; 31: 1889–1896.
Mukherjee B, McEllin B, Camacho CV, Tomimatsu N, Sirasanagandala S, Nannepaga S et al EGFRvIII and DNA double-strand break repair: a molecular mechanism for radioresistance in glioblastoma. Cancer Res 2009; 69: 4252–4259.
Sutter R, Yadirgi G, Marino S . Neural stem cells, tumour stem cells and brain tumours: dangerous relationships? Biochim Biophys Acta 2007; 1776: 125–137.
Hambardzumyan D, Parada LF, Holland EC, Charest A . Genetic modeling of gliomas in mice: new tools to tackle old problems. Glia 2011; 59: 1155–1168.
Louis DN, Ohgaki H, Wiestler OD, Cavenee WK, Burger PC, Jouvet A et al The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol 2007; 114: 97–109.
Beroukhim R, Getz G, Nghiemphu L, Barretina J, Hsueh T, Linhart D et al Assessing the significance of chromosomal aberrations in cancer: methodology and application to glioma. Proc Natl Acad Sci USA 2007; 104: 20007–20012.
Fischer U, Muller HW, Sattler HP, Feiden K, Zang KD, Meese E . Amplification of the MET gene in glioma. Genes Chromosomes Cancer 1995; 12: 63–65.
Sottoriva A, Spiteri I, Piccirillo SG, Touloumis A, Collins VP, Marioni JC et al Intratumor heterogeneity in human glioblastoma reflects cancer evolutionary dynamics. Proc Natl Acad Sci USA. 2013; 110: 4009–4014.
Smolen GA, Muir B, Mohapatra G, Barmettler A, Kim WJ, Rivera MN et al Frequent met oncogene amplification in a Brca1/Trp53 mouse model of mammary tumorigenesis. Cancer Res 2006; 66: 3452–3455.
Bigner SH, Humphrey PA, Wong AJ, Vogelstein B, Mark J, Friedman HS et al Characterization of the epidermal growth factor receptor in human glioma cell lines and xenografts. Cancer Res 1990; 50: 8017–8022.
Li Y, Li A, Glas M, Lal B, Ying M, Sang Y et al c-Met signaling induces a reprogramming network and supports the glioblastoma stem-like phenotype. Proc Natl Acad Sci USA. 2011; 108: 9951–9956.
McLendon R, Friedman A, Bigner D, Van Meir EG, Brat DJ, Mastrogianakis M et alComprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature 2008; 455: 1061–1068.
Attolini CS, Cheng YK, Beroukhim R, Getz G, Abdel-Wahab O, Levine RL et al A mathematical framework to determine the temporal sequence of somatic genetic events in cancer. Proc Natl Acad Sci USA. 2010; 107: 17604–17609.
Mukherjee B, Camacho CV, Tomimatsu N, Miller J, Burma S . Modulation of the DNA-damage response to HZE particles by shielding. DNA Repair (Amst) 2008; 7: 1717–1730.
Costes SV, Boissiere A, Ravani S, Romano R, Parvin B, Barcellos-Hoff MH . Imaging features that discriminate between foci induced by high- and low-LET radiation in human fibroblasts. Radiat Res 2006; 165: 505–515.
Saha J, Wang M, Cucinotta FA . Investigation of switch from ATM to ATR signaling at the sites of DNA damage induced by low and high LET radiation. DNA Repair (Amst) 2013; 12: 1143–1151.
Wang H, Wang X, Zhang P, Wang Y . The Ku-dependent non-homologous end-joining but not other repair pathway is inhibited by high linear energy transfer ionizing radiation. DNA Repair (Amst) 2008; 7: 725–733.
Yajima H, Fujisawa H, Nakajima NI, Hirakawa H, Jeggo PA, Okayasu R et al The complexity of DNA double strand breaks is a critical factor enhancing end-resection. DNA Repair (Amst) 2013; 12: 936–946.
Krimpenfort P, Ijpenberg A, Song JY, van der Valk M, Nawijn M, Zevenhoven J et al p15Ink4b is a critical tumour suppressor in the absence of p16Ink4a. Nature 2007; 448: 943–946.
Trusolino L, Bertotti A, Comoglio PM . MET signalling: principles and functions in development, organ regeneration and cancer. Nat Rev Mol Cell Biol 2010; 11: 834–848.
Gherardi E, Birchmeier W, Birchmeier C, Vande Woude G . Targeting MET in cancer: rationale and progress. Nat Rev Cancer 2012; 12: 89–103.
De Bacco F, Casanova E, Medico E, Pellegatta S, Orzan F, Albano R et al The MET oncogene is a functional marker of a glioblastoma stem cell subtype. Cancer Res 2012; 72: 4537–4550.
Joo KM, Jin J, Kim E, Ho Kim K, Kim Y, Gu Kang B et al MET signaling regulates glioblastoma stem cells. Cancer Res 2012; 72: 3828–3838.
Hellman A, Zlotorynski E, Scherer SW, Cheung J, Vincent JB, Smith DI et al A role for common fragile site induction in amplification of human oncogenes. Cancer Cell 2002; 1: 89–97.
Li Y, Lal B, Kwon S, Fan X, Saldanha U, Reznik TE et al The scatter factor/hepatocyte growth factor: c-met pathway in human embryonal central nervous system tumor malignancy. Cancer Res 2005; 65: 9355–9362.
Laterra J, Rosen E, Nam M, Ranganathan S, Fielding K, Johnston P . Scatter factor/hepatocyte growth factor expression enhances human glioblastoma tumorigenicity and growth. Biochem Biophys Res Commun 1997; 235: 743–747.
Engelman JA, Zejnullahu K, Mitsudomi T, Song Y, Hyland C, Park JO et al MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science 2007; 316: 1039–1043.
Jun HJ, Acquaviva J, Chi D, Lessard J, Zhu H, Woolfenden S et al Acquired MET expression confers resistance to EGFR inhibition in a mouse model of glioblastoma multiforme. Oncogene 2012; 31: 3039–3050.
Turke AB, Zejnullahu K, Wu YL, Song Y, Dias-Santagata D, Lifshits E et al Preexistence and clonal selection of MET amplification in EGFR mutant NSCLC. Cancer Cell 2010; 17: 77–88.
Stommel JM, Kimmelman AC, Ying H, Nabioullin R, Ponugoti AH, Wiedemeyer R et al Coactivation of receptor tyrosine kinases affects the response of tumor cells to targeted therapies. Science 2007; 318: 287–290.
De Bacco F, Luraghi P, Medico E, Reato G, Girolami F, Perera T et al Induction of MET by ionizing radiation and its role in radioresistance and invasive growth of cancer. J Natl Cancer Inst 2011; 103: 645–661.
Liu W, Fu Y, Xu S, Ding F, Zhao G, Zhang K et al c-Met expression is associated with time to recurrence in patients with glioblastoma multiforme. J Clin Neurosci 2011; 18: 119–121.
Isaka F, Ishibashi M, Taki W, Hashimoto N, Nakanishi S, Kageyama R . Ectopic expression of the bHLH gene Math1 disturbs neural development. Eur J Neurosci 1999; 11: 2582–2588.
Krimpenfort P, Quon KC, Mooi WJ, Loonstra A, Berns A . Loss of p16Ink4a confers susceptibility to metastatic melanoma in mice. Nature 2001; 413: 83–86.
Podsypanina K, Ellenson LH, Nemes A, Gu J, Tamura M, Yamada KM et al Mutation of Pten/Mmac1 in mice causes neoplasia in multiple organ systems. Proc Natl Acad Sci USA 1999; 96: 1563–1568.
Scotto L, Narayan G, Nandula SV, Subramaniyam S, Kaufmann AM, Wright JD et al Integrative genomics analysis of chromosome 5p gain in cervical cancer reveals target over-expressed genes, including Drosha. Mol Cancer 2008; 7: 58.
Acknowledgements
SB is supported by grants from the National Aeronautics and Space Administration (NNX13AI13G), National Institutes of Health (RO1 CA149461) and the Cancer Prevention and Research Institute of Texas (RP100644). AAH is supported by a National Institutes of Health grant (R01 NS062080). MCH is supported by a National Institute of General Medical Sciences training grant 5T32GM008203 in cellular and molecular biology. CVC was supported by a NCI training grant (T32CA124334). CVC completed this work in partial fulfillment of the requirements for her PhD degree. We thank Dr Chaitanya Nirodi for valuable advice on cloning and lentivirus production strategies. We also thank the support staff at the NASA Space Radiation Research Laboratory, Brookhaven National Laboratory, Upton, NY, USA, for facilitating particle radiation experiments.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Oncogene website
Supplementary information
Rights and permissions
About this article
Cite this article
Camacho, C., Todorova, P., Hardebeck, M. et al. DNA double-strand breaks cooperate with loss of Ink4 and Arf tumor suppressors to generate glioblastomas with frequent Met amplification. Oncogene 34, 1064–1072 (2015). https://doi.org/10.1038/onc.2014.29
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/onc.2014.29
This article is cited by
-
The DNA Double-Strand Break Repair in Glioma: Molecular Players and Therapeutic Strategies
Molecular Neurobiology (2022)
-
MET in glioma: signaling pathways and targeted therapies
Journal of Experimental & Clinical Cancer Research (2019)
-
The genetic landscape of gliomas arising after therapeutic radiation
Acta Neuropathologica (2019)
-
Phytosomal curcumin causes natural killer cell-dependent repolarization of glioblastoma (GBM) tumor-associated microglia/macrophages and elimination of GBM and GBM stem cells
Journal of Experimental & Clinical Cancer Research (2018)
-
DNA damage interactions on both nanometer and micrometer scale determine overall cellular damage
Scientific Reports (2018)