Abstract
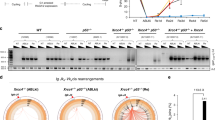
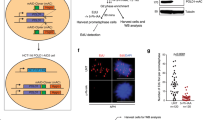
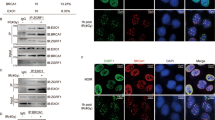
MLL (myeloid/lymphoid or mixed-lineage leukemia) rearrangements are frequent in therapy-related and childhood acute leukemia, and are associated with poor prognosis. The majority of the rearrangements fall within a 7.3-kb MLL breakpoint cluster region (MLLbcr), particularly in a 0.4-kb hotspot at the intron11-exon12 boundary. The underlying mechanisms are poorly understood, though multiple pathways including early apoptotic signaling, accompanied by high-order DNA fragmentation, have been implicated. We introduced the MLLbcr hotspot in an EGFP-based recombination reporter system and demonstrated enhancement of both spontaneous and genotoxic treatment-induced DNA recombination by the MLLbcr in various human cell types. We identified Endonuclease G (EndoG), an apoptotic nuclease, as an essential factor for MLLbcr-specific DNA recombination after induction of replication stress. We provide evidence for replication stress-induced nuclear accumulation of EndoG, DNA binding by EndoG as well as cleavage of the chromosomal MLLbcr locus in a manner requiring EndoG. We demonstrate additional dependency of MLLbcr breakage on ATM signaling to histone H2B monoubiquitinase RNF20, involved in chromatin relaxation. Altogether our findings provide a novel mechanism underlying MLLbcr destabilization in the cells of origin of leukemogenesis, with replication stress-activated, EndoG-mediated cleavage at the MLLbcr, which may serve resolution of the stalled forks via recombination repair, however, also permits MLL rearrangements.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Mosad E, Abdou M, Zaky AH . Rearrangement of the myeloid/lymphoid leukemia gene in therapy-related myelodysplastic syndrome in patients previously treated with agents targeting DNA topoisomerase II. Oncology 2012; 83: 128–134.
Abdulwahab A, Sykes J, Kamel-Reid S, Chang H, Brandwein JM . Therapy-related acute lymphoblastic leukemia is more frequent than previously recognized and has a poor prognosis. Cancer 2012; 118: 3962–3967.
Stam RW, den Boer ML, Pieters R . Towards targeted therapy for infant acute lymphoblastic leukemia. Br J Haematol 2006; 132: 539–551.
Shivakumar R, Tan W, Wilding GE, Wang ES, Wetzler M . Biologic features and treatment outcome of secondary acute lymphoblastic leukemia—a review of 101 cases. Ann Oncol 2008; 19: 1634–1638.
Ezoe S . Secondary leukemia associated with the anti-cancer agent, etoposide, a topoisomerase II inhibitor. Int J Environ Res Public Health 2012; 9: 2444–2453.
Meyer C, Hofmann J, Burmeister T, Gröger D, Park TS, Emerenciano M et al. The MLL recombinome of acute leukemias in 2013. Leukemia 2013; 27: 2165–2176.
Stanulla M, Wang J, Chervinsky DS, Thandla S, Aplan PD . DNA cleavage within the MLL breakpoint cluster region is a specific event which occurs as part of higher-order chromatin fragmentation during the initial stages of apoptosis. Mol Cell Biol 1997; 17: 4070–4079.
Mirault ME, Boucher P, Tremblay A . Nucleotide-resolution mapping of topoisomerase-mediated and apoptotic DNA strand scissions at or near an MLL translocation hotspot. Am J Hum Genet 2006; 79: 779–791.
Sim SP, Liu LF . Nucleolytic cleavage of the mixed lineage leukemia breakpoint cluster region during apoptosis. J Biol Chem 2001; 276: 31590–31595.
Hars ES, Lyu YL, Lin CP, Liu LF . Role of apoptotic nuclease caspase-activated DNase in etoposide-induced treatment-related acute myelogenous leukemia. Cancer Res 2006; 66: 8975–8979.
Samejima K, Earnshaw WC . Trashing the genome: the role of nucleases during apoptosis. Nat Rev Mol Cell Biol 2005; 6: 677–688.
Britton S, Frit P, Biard D, Salles B, Calsou P . ARTEMIS nuclease facilitates apoptotic chromatin cleavage. Cancer Res 2009; 69: 8120–8126.
Libura J, Ward M, Solecka J, Richardson C . Etoposide-initiated MLL rearrangements detected at high frequency in human primitive hematopoietic stem cells with in vitro and in vivo long-term repopulating potential. Eur J Haematol 2008; 81: 185–195.
Stanulla M, Chhalliyil P, Wang J, Jani-Sait SN, Aplan PD . Mechanisms of MLL gene rearrangement: site-specific DNA cleavage within the breakpoint cluster region is independent of chromosomal context. Hum Mol Genet 2001; 10: 2481–2491.
Boehden GS, Restle A, Marschalek R, Stocking C, Wiesmüller L . Recombination at chromosomal sequences involved in leukaemogenic rearrangements is differentially regulated by p53. Carcinogenesis 2004; 25: 1305–1313.
Akyüz N, Boehden GS, Süsse S, Rimek A, Preuss U, Scheidtmann KH et al. DNA substrate dependence of p53-mediated regulation of double-strand break repair. Mol Cell Biol 2002; 22: 6306–6317.
Strissel PL, Strick R, Rowley JD, Zeleznik-Le NJ . An in vivo topoisomerase II cleavage site and a DNase I hypersensitive site colocalize near exon 9 in the MLL breakpoint cluster region. Blood 1998; 92: 3793–3803.
Flynn RL, Zou L . ATR: a master conductor of cellular responses to DNA replication stress. Trends Biochem Sci 2011; 36: 133–140.
Matsuoka S, Ballif BA, Smogorzewska A, McDonald ER 3rd, Hurov KE, Luo J et al. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science 2007; 316: 1160–1166.
Carrassa L, Broggini M, Erba E, Damia G . Chk1, but not Chk2, is involved in the cellular response to DNA damaging agents: differential activity in cells expressing or not p53. Cell Cycle 2004; 3: 1177–1181.
Vařecha M, Potěšilová M, Matula P, Kozubek M . Endonuclease G interacts with histone H2B and DNA topoisomerase II alpha during apoptosis. Mol Cell Biochem 2012; 363: 301–307.
Wu J, Huen MS, Lu LY, Ye L, Dou Y, Ljungman M et al. Histone ubiquitination associates with BRCA1-dependent DNA damage response. Mol Cell Biol 2009; 29: 849–860.
Moyal L, Lerenthal Y, Gana-Weisz M, Mass G, So S, Wang SY et al. Requirement of ATM-dependent monoubiquitylation of histone H2B for timely repair of DNA double-strand breaks. Mol Cell 2011; 41: 529–542.
Maddams J, Parkin DM, Darby SC . The cancer burden in the United Kingdom in 2007 due to radiotherapy. Int J Cancer 2011; 129: 2885–2893.
Betti CJ, Villalobos MJ, Diaz MO, Vaughan AT . Apoptotic stimuli initiate MLL-AF9 translocations that are transcribed in cells capable of division. Cancer Res 2003; 63: 1377–1381.
Saintigny Y, Delacôte F, Varès G, Petitot F, Lambert S, Averbeck D et al. Characterization of homologous recombination induced by replication inhibition in mammalian cells. EMBO J 2001; 20: 3861–3870.
Ozeri-Galai E, Schwartz M, Rahat A, Kerem B . Interplay between ATM and ATR in the regulation of common fragile site stability. Oncogene 2008; 27: 2109–2117.
Sordet O, Khan QA, Plo I, Pourquier P, Urasaki Y, Yoshida A et al. Apoptotic topoisomerase I-DNA complexes induced by staurosporine-mediated oxygen radicals. J Biol Chem 2004; 279: 50499–50504.
Li LY, Luo X, Wang X . Endonuclease G is an apoptotic DNase when released from mitochondria. Nature 2001; 412: 95–99.
Büttner S, Eisenberg T, Carmona-Gutierrez D, Ruli D, Knauer H, Ruckenstuhl C et al. Endonuclease G regulates budding yeast life and death. Mol Cell 2007; 25: 233–246.
Tichy ED, Stephan ZA, Osterburg A, Noel G, Stambrook PJ . Mouse embryonic stem cells undergo charontosis, a novel programmed cell death pathway dependent upon cathepsins, p53, and EndoG, in response to etoposide treatment. Stem Cell Res 2013; 10: 428–441.
Ruiz-Carrillo A, Renaud J . Endonuclease G: a (dG)n X (dC)n-specific DNase from higher eukaryotes. EMBO J 1987; 6: 401–407.
Diener T, Neuhaus M, Koziel R, Micutkova L, Jansen-Dürr P . Role of endonuclease G in senescence-associated cell death of human endothelial cells. Exp Gerontol 2010; 45: 638–644.
Yang SH, Chien CM, Lu MC, Lin YH, Hu XW, Lin SR . Up-regulation of Bax and endonuclease G, and down-modulation of Bcl-XL involved in cardiotoxin III-induced apoptosis in K562 cells. Exp Mol Med 2006; 38: 435–444.
Zan H, Zhang J, Al-Qahtani A, Pone EJ, White CA, Lee D et al. Endonuclease G plays a role in immunoglobulin class switch DNA recombination by introducing double-strand breaks in switch regions. Mol Immunol 2011; 48: 610–622.
Lee JS, Seo TW, Yi JH, Shin KS, Yoo SJ . CHIP has a protective role against oxidative stress-induced cell death through specific regulation of Endonuclease G. Cell Death Dis 2013; 4: e666.
Ozeri-Galai E, Lebofsky R, Rahat A, Bester AC, Bensimon A, Kerem B . Failure of origin activation in response to fork stalling leads to chromosomal instability at fragile sites. Mol Cell 2011; 43: 122–131.
Barlow JH, Faryabi RB, Callén E, Wong N, Malhowski A, Chen HT et al. Identification of early replicating fragile sites that contribute to genome instability. Cell 2013; 152: 620–632.
Bolderson E, Scorah J, Helleday T, Smythe C, Meuth M . ATM is required for the cellular response to thymidine induced replication fork stress. Hum Mol Genet 2004; 13: 2937–2945.
Ousset M, Bouquet F, Fallone F, Biard D, Dray C, Valet P et al. Loss of ATM positively regulates the expression of hypoxia inducible factor 1 (HIF-1) through oxidative stress: Role in the physiopathology of the disease. Cell Cycle 2010; 9: 2814–2822.
Sun J, Oma Y, Harata M, Kono K, Shima H, Kinomura A et al. ATM modulates the loading of recombination proteins onto a chromosomal translocation breakpoint hotspot. PLoS ONE 2010; 5: e13554.
Volčič M, Karl S, Baumann B, Salles D, Daniel P, Fulda S et al. NF-κB regulates DNA double-strand break repair in conjunction with BRCA1-CtIP complexes. Nucleic Acids Res 2012; 40: 181–195.
Grabarz A, Barascu A, Guirouilh-Barbat J, Lopez BS . Initiation of DNA double strand break repair: signaling and single-stranded resection dictate the choice between homologous recombination, non-homologous end-joining and alternative end-joining. Am J Cancer Res 2012; 2: 249–268.
Fierz B, Chatterjee C, McGinty RK, Bar-Dagan M, Raleigh DP, Muir TW . Histone H2B ubiquitylation disrupts local and higher-order chromatin compaction. Nat Chem Biol 2011; 7: 113–119.
Chernikova SB, Razorenova OV, Higgins JP, Sishc BJ, Nicolau M, Dorth JA et al. Deficiency in mammalian histone H2B ubiquitin ligase Bre1 (Rnf20/Rnf40) leads to replication stress and chromosomal instability. Cancer Res 2012; 72: 2111–2119.
Zhang F, Carvalho CM, Lupski JR . Complex human chromosomal and genomic rearrangements. Trends Genet 2009; 25: 298–307.
Milyavsky M, Gan OI, Trottier M, Komosa M, Tabach O, Notta F et al. A distinctive DNA damage response in human hematopoietic stem cells reveals an apoptosis-independent role for p53 in self-renewal. Cell Stem Cell 2010; 7: 186–197.
Labi V, Erlacher M, Krumschnabel G, Manzl C, Tzankov A, Pinon J et al. Apoptosis of leukocytes triggered by acute DNA damage promotes lymphoma formation. Genes Dev 2010; 24: 1602–1607.
Baumann C, Boehden GS, Bürkle A, Wiesmüller L . Poly(ADP-RIBOSE) polymerase-1 (Parp-1) antagonizes topoisomerase I-dependent recombination stimulation by P53. Nucleic Acids Res 2006; 34: 1036–1049.
Acknowledgements
We thank Daniela Waldraff, Regina Häfele, Rahel Wiehe (all Department of Obstetrics and Gynecology, University of Ulm, Germany) and Dré van der Merwe (Core facility FACS, Medical Faculty, University of Ulm, Germany) for their contributions to this work. This work was supported by the German Research Foundation (WI 3099/7-1, WI 3099/7-2) and by the European/German Space Agency (ESA/DLR) and German Ministry of Economy (BMWi), A0-10-IBER-2 funding 50WB1225.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Oncogene website
Supplementary information
Rights and permissions
About this article
Cite this article
Gole, B., Baumann, C., Mian, E. et al. Endonuclease G initiates DNA rearrangements at the MLL breakpoint cluster upon replication stress. Oncogene 34, 3391–3401 (2015). https://doi.org/10.1038/onc.2014.268
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/onc.2014.268
This article is cited by
-
Vpu modulates DNA repair to suppress innate sensing and hyper-integration of HIV-1
Nature Microbiology (2020)
-
Base excision repair proteins couple activation-induced cytidine deaminase and endonuclease G during replication stress-induced MLL destabilization
Leukemia (2018)
-
Executioner caspases and CAD are essential for mutagenesis induced by TRAIL or vincristine
Cell Death & Disease (2017)
-
NF-κB-dependent DNA damage-signaling differentially regulates DNA double-strand break repair mechanisms in immature and mature human hematopoietic cells
Leukemia (2015)