Abstract
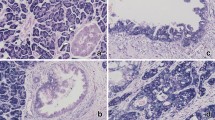
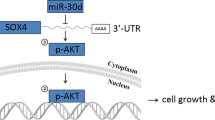
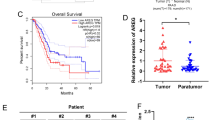
Pancreatic ductal adenocarcinoma (PDAC) is a highly invasive cancer with a poor prognosis. Although microRNA (miRNA) transcripts have a crucial role in carcinogenesis and development, little information is known regarding the aberrant DNA methylation of miRNAs in PDAC. Using methylated DNA immunoprecipitation-chip analysis, we found that miR-615-5p was hypermethylated in its putative promoter region, which silenced its expression in PDAC cell lines. In addition, the overexpression of miR-615-5p in pancreatic cancer cells suppressed cell proliferation, migration and invasion. Insulin-like growth factor 2 (IGF2) is an imprinted gene, and its abnormal expression contributes to tumor growth. Here, we identified IGF2 as a target of miR-615-5p using a luciferase reporter assay. IGF2 upregulation in PDAC tissues was not correlated with a loss of imprinting but was inversely correlated with miR-615-5p downregulation. In addition, miR-615-5p suppressed pancreatic cancer cell proliferation, migration and invasion by directly targeting IGF2, and this effect could be reversed by co-transfection with IGF2. Furthermore, the stable overexpression of miR-615-5p inhibited tumor growth in vivo and was correlated with IGF2 expression. Using RNA sequencing, we further identified miR-615-5p as potentially targeting other genes, such as the proto-oncogene JUNB, and interfering with the insulin signaling pathway. Taken together, our results demonstrate that miR-615-5p was abnormally downregulated in PDAC cells due to promoter hypermethylation, which limited its inhibition of IGF2 and other target genes, thereby contributing to tumor growth, invasion and migration. These data demonstrate a novel and important role of miR-615-5p as a tumor suppressor in PDAC.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Accession codes
References
Siegel R, Ward E, Brawley O, Jemal A Cancer statistics, 2011: the impact of eliminating socioeconomic and racial disparities on premature cancer deaths. CA Cancer J Clin 2011; 61: 212–236.
Egger G, Liang G, Aparicio A, Jones PA Epigenetics in human disease and prospects for epigenetic therapy. Nature 2004; 429: 457–463.
Kim M, Kasinski AL, Slack FJ MicroRNA therapeutics in preclinical cancer models. Lancet Oncol 2011; 12: 319–321.
Png KJ, Halberg N, Yoshida M, Tavazoie SF A microRNA regulon that mediates endothelial recruitment and metastasis by cancer cells. Nature 2012; 481: 190–194.
Boeri M, Verri C, Conte D, Roz L, Modena P, Facchinetti F et al. MicroRNA signatures in tissues and plasma predict development and prognosis of computed tomography detected lung cancer. Proc Natl Acad Sci USA 2011; 108: 3713–3718.
van Kouwenhove M, Kedde M, Agami R MicroRNA regulation by RNA-binding proteins and its implications for cancer. Nat Rev Cancer 2011; 11: 644–656.
Wang J, Sen S MicroRNA functional network in pancreatic cancer: from biology to biomarkers of disease. J Biosci 2011; 36: 481–491.
De Carvalho DD, Sharma S, You JS, Su SF, Taberlay PC, Kelly TK et al. DNA methylation screening identifies driver epigenetic events of cancer cell survival. Cancer Cell 2012; 21: 655–667.
Teschendorff AE, Widschwendter M Differential variability improves the identification of cancer risk markers in DNA methylation studies profiling precursor cancer lesions. Bioinformatics 2012; 28: 1487–1494.
Han L, Witmer PD, Casey E, Valle D, Sukumar S DNA methylation regulates microRNA expression. Cancer Biol Ther 2007; 6: 1284–1288.
Kunej T, Godnic I, Ferdin J, Horvat S, Dovc P, Calin GA Epigenetic regulation of microRNAs in cancer: an integrated review of literature. Mutat Res 2011; 717: 77–84.
Lujambio A, Ropero S, Ballestar E, Fraga MF, Cerrato C, Setien F et al. Genetic unmasking of an epigenetically silenced microRNA in human cancer cells. Cancer Res 2007; 67: 1424–1429.
Weber B, Stresemann C, Brueckner B, Lyko F Methylation of human microRNA genes in normal and neoplastic cells. Cell Cycle 2007; 6: 1001–1005.
Weber M, Davies JJ, Wittig D, Oakeley EJ, Haase M, Lam WL et al. Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells. Nat Genet 2005; 37: 853–862.
Gao W, Kondo Y, Shen L, Shimizu Y, Sano T, Yamao K et al. Variable DNA methylation patterns associated with progression of disease in hepatocellular carcinomas. Carcinogenesis 2008; 29: 1901–1910.
Soares MR, Huber J, Rios AF, Ramos ES Investigation of IGF2/ApaI and H19/RsaI polymorphisms in patients with cutaneous melanoma. Growth Horm IGF Res 2010; 20: 295–297.
Kondo Y, Kanai Y, Sakamoto M, Mizokami M, Ueda R, Hirohashi S Genetic instability and aberrant DNA methylation in chronic hepatitis and cirrhosis—a comprehensive study of loss of heterozygosity and microsatellite instability at 39 loci and DNA hypermethylation on 8 CpG islands in microdissected specimens from patients with hepatocellular carcinoma. Hepatology 2000; 32: 970–979.
Mardin WA, Mees ST MicroRNAs: novel diagnostic and therapeutic tools for pancreatic ductal adenocarcinoma? Ann Surg Oncol 2009; 16: 3183–3189.
Rachagani S, Kumar S, Batra SK MicroRNA in pancreatic cancer: pathological, diagnostic and therapeutic implications. Cancer Lett 2010; 292: 8–16.
Fiorucci G, Chiantore MV, Mangino G, Percario ZA, Affabris E, Romeo G Cancer regulator microRNA: potential relevance in diagnosis, prognosis and treatment of cancer. Curr Med Chem 2012; 19: 461–474.
Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D et al. MicroRNA expression profiles classify human cancers. Nature 2005; 435: 834–838.
Tricoli JV, Jacobson JW MicroRNA: potential for cancer detection, diagnosis, and prognosis. Cancer Res 2007; 67: 4553–4555.
Saito Y, Liang G, Egger G, Friedman JM, Chuang JC, Coetzee GA et al. Specific activation of microRNA-127 with downregulation of the proto-oncogene BCL6 by chromatin-modifying drugs in human cancer cells. Cancer Cell 2006; 9: 435–443.
Sato F, Tsuchiya S, Meltzer SJ, Shimizu K MicroRNAs and epigenetics. FEBS J 2011; 278: 1598–1609.
Zhou X, Ruan J, Wang G, Zhang W Characterization and identification of microRNA core promoters in four model species. PLoS Comput Biol 2007; 3: e37.
Chao W, D'Amore PA IGF2: epigenetic regulation and role in development and disease. Cytokine Growth Factor Rev 2008; 19: 111–120.
Dong X, Li Y, Tang H, Chang P, Hess KR, Abbruzzese JL et al. Insulin-like growth factor axis gene polymorphisms modify risk of pancreatic cancer. Cancer Epidemiol 2012; 36: 206–211.
Suzuki H, Li Y, Dong X, Hassan MM, Abbruzzese JL, Li D Effect of insulin-like growth factor gene polymorphisms alone or in interaction with diabetes on the risk of pancreatic cancer. Cancer Epidemiol Biomarkers Prev 2008; 17: 3467–3473.
Hahn H, Wojnowski L, Specht K, Kappler R, Calzada-Wack J, Potter D et al. Patched target Igf2 is indispensable for the formation of medulloblastoma and rhabdomyosarcoma. J Biol Chem 2000; 275: 28341–28344.
Ingram WJ, Wicking CA, Grimmond SM, Forrest AR, Wainwright BJ Novel genes regulated by Sonic Hedgehog in pluripotent mesenchymal cells. Oncogene 2002; 21: 8196–8205.
El TH, Hosny KA, Esmat G, Breuhahn K, Abdelaziz AI miR-615-5p is restrictedly expressed in cirrhotic and cancerous liver tissues and its overexpression alleviates the tumorigenic effects in hepatocellular carcinoma. FEBS Lett 2012; 586: 3309–3316.
Yang Y, Wang LL, Li YH, Gao XN, Liu Y, Yu L Effect of CpG island methylation on microRNA expression in the k-562 cell line. Biochem Genet 2012; 50: 122–134.
Jacinto FV, Ballestar E, Esteller M Methyl-DNA immunoprecipitation (MeDIP): hunting down the DNA methylome. Biotechniques 2008; 44: 37–43.
Cui H, Horon IL, Ohlsson R, Hamilton SR, Feinberg AP Loss of imprinting in normal tissue of colorectal cancer patients with microsatellite instability. Nat Med 1998; 4: 1276–1280.
Rainier S, Johnson LA, Dobry CJ, Ping AJ, Grundy PE, Feinberg AP . Relaxation of imprinted genes in human cancer. Nature 1993; 362: 747–749.
Mou HQ, Lu J, Zhu SF, Lin CL, Tian GZ, Xu X et al. Transcriptomic analysis of paulownia infected by paulownia witches'-broom phytoplasma. PLoS ONE 2013; 8: e77217.
Acknowledgements
This work was supported by grants from the National Nature Science Foundation of China (Nos 81172267 and 30901627), the National Project For Translational Research Of Early Diagnosis And Comprehensive Treatment In Pancreatic Cancer (201202007) and the Priority Academic Program Development of Jiangsu Higher Education Institutions (JX10231801).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Oncogene website
Rights and permissions
About this article
Cite this article
Gao, W., Gu, Y., Li, Z. et al. miR-615-5p is epigenetically inactivated and functions as a tumor suppressor in pancreatic ductal adenocarcinoma. Oncogene 34, 1629–1640 (2015). https://doi.org/10.1038/onc.2014.101
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/onc.2014.101
This article is cited by
-
The downregulation of miR-509-3p expression by collagen type XI alpha 1-regulated hypermethylation facilitates cancer progression and chemoresistance via the DNA methyltransferase 1/Small ubiquitin-like modifier-3 axis in ovarian cancer cells
Journal of Ovarian Research (2023)
-
IGF2 is upregulated by its antisense RNA to potentiate pancreatic cancer progression
Functional & Integrative Genomics (2023)
-
Iodine-125 seed represses the growth and facilitates the apoptosis of colorectal cancer cells by suppressing the methylation of miR-615 promoter
BMC Cancer (2022)
-
LncRNA LINC00461 exacerbates myocardial ischemia–reperfusion injury via microRNA-185-3p/Myd88
Molecular Medicine (2022)
-
circSETD3 regulates MAPRE1 through miR-615-5p and miR-1538 sponges to promote migration and invasion in nasopharyngeal carcinoma
Oncogene (2021)