Abstract
Chronic inflammation is an established risk factor for the onset of cancer, and the inflammatory cytokine IL-6 has a role in tumorigenesis by enhancing proliferation and hindering apoptosis. As factors stimulating proliferation also downregulate p53 expression by enhancing ribosome biogenesis, we hypothesized that IL-6 may cause similar changes in inflamed tissues, thus activating a mechanism that favors neoplastic transformation. Here, we showed that IL-6 downregulated the expression and activity of p53 in transformed and untransformed human cell lines. This was the consequence of IL-6-dependent stimulation of c-MYC mRNA translation, which was responsible for the upregulation of rRNA transcription. The enhanced rRNA transcription stimulated the MDM2-mediated proteasomal degradation of p53, by reducing the availability of ribosome proteins for MDM2 binding. The p53 downregulation induced the acquisition of cellular phenotypic changes characteristic of epithelial–mesenchymal transition, such as a reduced level of E-cadherin expression, increased cell invasiveness and a decreased response to cytotoxic stresses. We found that these changes also occurred in colon epithelial cells of patients with ulcerative colitis, a very representative example of chronic inflammation at high risk for tumor development. Histochemical and immunohistochemical analysis of colon biopsy samples showed an upregulation of ribosome biogenesis, a reduced expression of p53, together with a focal reduction or absence of E-cadherin expression in chronic colitis in comparison with normal mucosa samples. These changes disappeared after treatment with anti-inflammatory drugs. Taken together, the present results highlight a new mechanism that may link chronic inflammation to cancer, based on p53 downregulation, which is activated by the enhancement of rRNA transcription upon IL-6 exposure.
Similar content being viewed by others
Introduction
Chronic inflammation is an established risk factor for the development of many types of human cancers.1, 2 The inflammatory substances released in the inflamed tissue can contribute to all steps of carcinogenesis by favoring both tumor initiation, by inducing DNA damages involving oncogenes and/or tumor suppressor genes, and tumor promotion, by increasing proliferation and resistance to the apoptosis of mutated cells.3, 4 Among the various substances present in the inflammatory milieu, interleukin (IL)-6 has been proven to have a fundamental role in experimental colon and liver tumorigenesis.5, 6, 7, 8 Furthermore, the high plasma level of IL-6 is a major risk factor for developing human colorectal tumors9 and hepatocellular carcinomas.10 This close link between IL-6 and cancer suggests that the IL-6 action in enhancing cell proliferation and reducing apoptosis might not be the only tumor-inducing mechanism controlled by the cytokine, but that additional IL-6 effects that may be conducive to tumorigenesis should be postulated. We recently demonstrated that the upregulation of rRNA transcription by factors stimulating proliferation downregulates p53 expression and activity by enhancing the MDM2-mediated p53 proteasomal digestion.11 We hypothesized that in human pathological conditions in which cell proliferation stimulating factors are produced, such as those characterizing the chronic inflammation processes, a similar mechanism may be active, thus contributing to neoplastic transformation. Therefore, in the present study we first wondered whether IL-6 might stimulate rRNA transcription and downregulate p53 expression and activity in human transformed and untransformed cell lines. We found that IL-6 enhanced rRNA transcription and that the upregulated rRNA synthesis reduced p53 expression and function through the activation of the ribonucleoprotein-MDM2 degradation pathway of p53.12, 13 Moreover, we observed that the p53 downregulation was responsible for the acquisition by the IL-6-exposed cells of the phenotype and functional changes characteristic of epithelial–mesenchymal transition (EMT). Considering that the onset of colon cancer in patients with inflammatory bowel diseases is a very representative example of chronic inflammation-related tumorigenesis,14 we investigated whether the changes induced by IL-6 in the human cell lines were also present in the epithelial cells of human colonic mucosa with chronic ulcerative disease. We actually found that a marked nucleolar hypertrophy—a morphological sign of upregulated rRNA transcription15—was present in the epithelial cells of all cases of ulcerative colitis (UC) examined, together with a reduction in the p53 immunostaining. Moreover, the major EMT phenotypic change, the reduction of E-cadherin expression, also focally occurred in the epithelial cells of the colon mucosa of patients with UC. These data highlight a new pathway, activated by the IL-6-induced stimulation of rRNA transcription, linking inflammation to cancer.
Results
IL-6 stimulates rRNA transcription
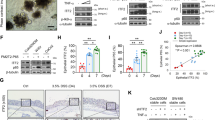
We analyzed the effect of IL-6 on ribosome biogenesis in the following four human epithelial cell lines: one from normal colon epithelium (NCM460 cell line), one from colon carcinoma (LS174T cell line), one from hepatocellular carcinoma (HepG2 cell line) and one from pancreas carcinoma (SW1990 cell line). The changes in rRNA transcriptional activity in these four cell lines exposed to IL-6 were ascertained using real-time–PCR analysis of the 45S prerRNA expression (Figure 1a). The rRNA synthesis was also evaluated by the immunofluorescence detection of nucleolar 5-fluorouridine (5-FU) incorporation into nascent rRNA in NCM460 and HepG2 cell lines (Figure 1b). Both methodological approaches revealed that IL-6 greatly enhanced rRNA transcription. As these data indicate that IL-6 enhances c-myc expression16, 17 in order to study the mechanism involved in the IL-6 stimulation of rRNA transcription, we investigated whether IL-6 enhanced the expression of c-myc protein also in the human epithelial cells used in the present study. For this reason we exposed both the NCM460 and the HepG2 cell lines to IL-6. We found that IL-6 significantly increased c-myc protein expression as early as 1 h after IL-6 stimulation in both cell lines, as evaluated by western blot analysis (Figure 1c). Unlike c-myc protein expression, the level of c-MYC mRNA did not change for up to 3 h after IL-6 exposure (Figure 1d). This suggested that, in our experimental conditions, a post-transcriptional mechanism was activated by IL-6 and was responsible for the increase in the c-myc protein. IL-6 has been reported to control the c-myc protein level either by increasing STAT3-mediated mRNA c-MYC transcription16 or via a stimulatory effect on the c-MYC internal ribosome entry site.17 Therefore, we analyzed the effect of IL-6 exposure on c-MYC mRNA IRES-dependent translation. For this purpose, NCM460 and HepG2 cells were transfected with a bicistronic in vitro transcribed mRNA in which the c-MYC-IRES sequence was inserted between two reporter luciferase cistrons (Renilla and Firefly, Promega, Milan, Italy). Evaluation of the ratio between the Firefly and Renilla activities indicated the IRES-dependent translation rate. We found that IL-6 treatment strongly stimulated the translation initiation mediated by the c-MYC-IRES in NCM460 and HepG2 cells (Figure 1e). As a control, a similar bicistronic transcript not containing the c-MYC-IRES element was transfected in IL-6-treated and control cells. In this case, no differences in the ratio between the Firefly and Renilla activities were detected. These findings indicated that the increased expression of c-myc protein observed in both cell lines after IL-6 exposure was due to an enhanced c-MYC mRNA IRES-dependent translation. In order to exclude that other post-transcriptional mechanisms may be involved, such as changes in protein stability, we evaluated the c-myc protein half-life in IL-6-stimulated HepG2 cells after protein synthesis inhibition by cycloheximide at a dose capable of completely inhibiting protein synthesis. We found that the half-life of c-myc protein was unchanged after IL-6 treatment (Figure 1f).
IL-6 treatment stimulates rRNA transcription by activation of c-myc protein in NCM460 and HepG2 cell lines. (a) Real-time–PCR analysis of the 45S rRNA expression in NCM460, HepG2, SW1990 and LS174T cells after 24 h of IL-6 treatment performed with a dose of 50 ng/ml. (b) Visualization of rRNA synthesis in control and IL-6-treated NCM460 and HepG2 cells. Cells were labeled with 5-FU for 15 min, and 5-FU revealed by specific FITC-conjugated monoclonal antibody. DAPI counterstaining. Scale bar=20 μm. (c) Representative western blot and densitometric analysis of c-myc expression in NCM460 and HepG2 cells treated with IL-6 for 1 and 3 h. (d) Real-time–PCR analysis of the c-MYC mRNA levels in NCM460 and HepG2 after 1 and 3 h of IL-6 treatment. (e) IRES-mediated translation assessed by measuring the FLuc and RLuc activity in control and 4 h IL-6-treated NCM460 and HepG2 cells after 8 h transfection with a bicistronic mRNA transcribed either from pRF (top) or from pR-c-MYC-IRES-F (bottom). (f) Time-course analysis of c-myc protein expression in control and 24 h-IL-6-stimulated HepG2 cells, exposed to cycloheximide (CHX) at a concentration of 20 μg/ml. (g) Real-time–PCR and western blot analysis of c-MYC expression in scrambled control siRNA (SCR) and in 24 h c-MYC-silenced (MYC−) HepG2 cells exposed to IL-6 for 24 h. The right panel shows the 45S rRNA expression in the same experimental condition. Histograms show the values (mean±s.d.) of three experiments. *P<0.05; **P<0.01. n.s., not significant.
At this point, we wondered whether the stimulation of rRNA transcription was exclusively due to the IL-6-induced upregulation of c-myc protein expression. For this purpose, we downregulated the expression of the c-MYC mRNA by the small interference RNA procedure and evaluated the effect of IL-6 stimulation on the synthesis of rRNA in HepG2 cells. We found that a c-MYC RNA interference significantly reduced the expression of the c-MYC mRNA and c-myc protein and counteracted the stimulatory effect of IL-6 on rRNA synthesis (Figure 1g).
IL-6 downregulates p53 expression and activity
The inhibition of rRNA transcription allows a larger amount of ribosomal proteins, no longer used for ribosome building, to bind to MDM2, thus reducing the MDM2-mediated proteasomal degradation of p53 with consequent p53 stabilization.12, 13 Conversely, the upregulation of rRNA synthesis reduces the availability of ribosomal proteins for the binding to MDM2, thus increasing the MDM2-mediated proteasomal p53 digestion.11 Therefore, we wondered whether IL-6 would lower p53 expression and activity through the above-described mechanism. In fact, even though it has been reported that IL-6 activates STAT318 and that activated STAT3 binds to the TP53 gene promoter repressing the transcription of TP53 mRNA,19 we found that no significant change occurred in the transcription level of TP53 mRNA in the NCM460, HepG2, SW1990 and LS174T cell lines (Figure 2a). As western blot analysis demonstrated that IL-6 treatment actually reduced the amount of p53 in the NCM460, HepG2, SW1990 and LS174T cell lines (Figure 2b), we considered the possibility that in IL-6-stimulated cells the reduced availability of ribosomal proteins for MDM2 binding might be responsible for an increased p53 proteasomal degradation. For this reason, we evaluated the half-life of p53 by time-course western blot analysis in control and IL-6-stimulated HepG2 cells after treatment with cycloheximide. We found that the half-life of p53 in IL-6-stimulated cells was shorter than that of control cells (Figure 2c). Also, we treated IL-6-exposed HepG2 cells with the proteasome inhibitor MG-132. We found that the inhibition of proteasomal degradation canceled the difference between the p53 expression of control and stimulated cells without reducing the stimulation of rRNA transcription by IL-6 (Figure 2d). These data indicated that the downregulation of p53 expression was actually due to an increased protein degradation. The increased p53 degradation appeared to be the consequence of a reduced ribosomal protein binding to MDM2, which allowed MDM2 to bind a greater amount of p53 for digestion. In fact, coimmunoprecipitation analysis showed that the quantity of L11 ribosomal protein coimmunoprecipitated with MDM2 was reduced in IL-6-stimulated cells as compared with control cells, whereas the amount of p53 was increased (Figure 2e). In order to define the role of MDM2 in the reduction of p53 stabilization after IL-6 treatment, we evaluated the effect of the MDM2 inhibitor Nutlin-3 on p53 expression in control and IL-6-stimulated HepG2 cells. Nutlin-3 binds MDM2 in the p53-binding pocket, thus hindering the MDM2-mediated degradation of p53, which results in p53 stabilization and activation.20 Western blot analysis showed that Nutlin-3 induced a p53 stabilization that was quite similar in control and IL-6-stimulated cells (Figure 2f). This strongly suggested that the reduction of p53 expression after exposure to IL-6 was actually the consequence of an increased binding of MDM2 to p53. To exclude the possibility that IL-6 might mediate the effect on p53 independent of a reduction in ribosomal protein availability for MDM2 binding, we downregulated the expression of RPL11 by the small-interfering RNA procedure and evaluated the effect of IL-6 stimulation on p53 expression in HepG2 cells. We found that the reduction of RPL11 mRNA and RPL11 protein expression (Supplementary Figure 1) caused a strong reduction of p53 expression and that IL-6 treatment no longer reduced the expression of p53 in RPL11-silenced HepG2 cells (Figure 2g). This observation confirmed the importance of the reduction of ribosomal protein availability for MDM2 binding, in particular of RPL11, in the IL-6-induced p53 downregulation.
Stimulation of rRNA transcription by IL-6 downregulates p53 expression and activity. (a) Real-time–PCR evaluation of the TP53 mRNA expression in NCM460, HepG2, SW1990 and LS174T cells after 24 h of IL-6 treatment. (b) Representative western blot and densitometric analysis of p53 expression in NCM460, HepG2, SW1990 and LS174T cells treated with IL-6 for 24 h. (c) Representative western blot and time-course analysis of p53 protein expression in control and 24 h IL-6-stimulated HepG2 cells, exposed to cycloheximide (CHX) at a concentration of 20 μg/ml. The values relative to p53 expression at 0.5, 1 and 2 h of CHX treatment are significantly higher in control than in IL-6-exposed cells (P<0.01). (d) Western blot evaluation of p53 protein expression and real-time–PCR analysis of 45S rRNA in control and IL-6-treated HepG2 cells exposed to the proteasomal inhibitor MG-132 at a concentration of 10 μm for 2 h (e) Coimmunoprecipitation and densitometric analysis of the amount of p53 and RPL11 protein bound to MDM2 in control and IL-6-stimulated HepG2 cells. The amount of MDM2, p53 and L11 is shown in the first two lanes before immunoprecipitation (input). The third and fourth lanes show the quantity of non-immunoprecipitated proteins. The fifth and sixth lanes show the amount of MDM2, p53 and L11 proteins after immunoprecipitation with anti-MDM2 polyclonal antibody (IP:MDM2). (f) Representative western blot and densitometric analysis of p53 protein expression in control and 24 h IL-6-stimulated HepG2 cells treated with Nutlin-3 at a concentration of 5 mM for 16 h. (g) Representative western blot analysis of p53 expression in control (SCR), RPL11-silenced (L11−), MYC-silenced (MYC−) and RPL11− and MYC-silenced HepG2 cells. The cells were exposed, 48 h after the end of the silencing procedure, to IL-6 for 24 h. (h) Real-time–PCR analysis of the mRNA expression of p53 target genes BAX and PUMA in NCM460 and HepG2 cells exposed to IL-6 for 24 h. (i) Representative western blot of p53 expression in NCM460 and HepG2 cells exposed to IL-6 for 24 h and treated with 3.4 μM hydroxyurea or 20 μg/ml 5-FU for 4 h. Histograms show the values (mean±s.d.) of three experiments. *P<0.05; **P<0.01; n.s., not significant.
All together, these results strongly suggested that the reduction of p53 expression after IL-6 treatment was the consequence of the interleukin-induced increased translation of c-myc protein, which in turn, by enhancing rRNA transcription, reduced the inactivation of MDM2 by RPL11, thus favoring p53 protein digestion. In order to demonstrate that this was the case, we also evaluated the effect of IL-6 exposure on p53 expression in c-MYC-silenced HepG2 cells: we found that c-MYC RNA interference induced a significant p53 stabilization that was not quantitatively changed after IL-6 stimulation. In addition, depletion of RPL11 mRNA in c-MYC-silenced cells resulted in a strong reduction of p53 protein expression that, once again, was quite similar after IL-6 exposure (Figure 2g).
We also investigated whether the activity of p53 was reduced after IL-6 exposure. We measured the expression of two p53 target genes whose products have a tumor suppressor function: BAX and PUMA.21 The expression of p21waf1/cip1 was not evaluated because IL-6 exposure per se enhances p21 expression.22 The quantitative analysis of the mRNA expression of these two genes indicated that BAX and PUMA mRNAs were significantly reduced in NCM460 and HepG2 cells treated with IL-6 (Figure 2h). At this point, we wondered whether the reduced p53 level in cells with increased rRNA synthesis might also be reflected in a reduced p53 stabilization and activity after cytotoxic stress. For this purpose we used NCM460 and HepG2 cells exposed to IL-6 and treated with either Hydroxyurea or 5-FU for 4 h. Both drugs were responsible for p53 stabilization in NCM460 and HepG2 cells, and IL-6 stimulation caused a reduction in the level of stabilized p53 in NCM460 and HepG2 cells treated with hydroxyurea and 5-FU in comparison with unstimulated, drug-treated cells (Figure 2i) (see densitometric analysis in Supplementary Figure 2). In the case of 5-FU treatment, a longer exposure to the drug (12 h) caused a strong stabilization of p53 also in IL-6-stimulated cells. However, after 5-FU, the p53 amount in IL-6-treated cells was always lower in comparison with that in unstimulated cells. This may be because 5-FU did not alter rRNA transcription, which even after 12 h of drug exposure was higher in IL-6-stimulated than in control cells (see Supplementary Figure 3 and relative discussion ).
Regarding the expression of the p53 target genes, we observed that the BAX and PUMA mRNA level was markedly lower in IL-6-stimulated than in unstimulated NCM460 and HepG2 cells after either hydroxyurea or 5-FU treatment (Supplementary Figure 4). Therefore, these results indicated that IL-6 induced a lowering of the p53 level, which was responsible for the downregulation of the p53 target gene response, also after exposure to cytotoxic stresses.
IL-6 induces EMT by downregulating p53 expression
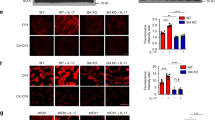
Downregulation of E-cadherin expression occurs very frequently during the progression of malignant epithelial tumors,23 and the loss of E-cadherin expression is responsible for the loss of intercellular adhesion during invasion.24 The E-cadherin expression is repressed by the transcription factor SLUG, responsible for EMT.25 As p53 controls the MDM2-mediated degradation of SLUG, 26 we wondered whether IL-6 could induce a reduction of E-cadherin expression by increasing the level of SLUG in a p53-dependent manner. For this purpose, we analyzed E-cadherin and SLUG protein expression: we found that at 24 h IL-6 stimulation increased the level of SLUG and reduced the expression of E-cadherin in NCM460 and HepG2 cell lines (Figure 3a). We did not observe any significant variation in the expression of E-cadherin and SLUG in TP53-silenced NCM460 cells (see the efficacy of TP53 silencing in Supplementary Figure 5).On the other hand, to avoid any misinterpretation on the role of p53 in reducing the level of E-cadherin in IL-6-treated cells, we used the HCT116 p53−/− cell line and analyzed the level of both E-cadherin and SLUG after IL-6 treatment. As shown in Figure 3a, IL-6 treatment did not modify the amount of both these proteins in HCT116 p53−/− cells, thus indicating the crucial role of p53 in this mechanism.
IL-6 induces EMT in a p53-dependent manner. (a) Representative western blot analysis of E-cadherin and SLUG expression in NCM460, HepG2 and HCT116 p53−/− cells exposed to IL-6 for 24 h. NCM460 were either (p53−) or not (SCR) silenced for TP53 expression. (b) Visualization of SLUG and E-cadherin distribution in control and IL-6-treated NCM460 cells. Cells were labeled with monoclonal antibodies versus SLUG or E-cadherin; the antibodies were revealed by FITC-conjugated secondary antibodies. Nuclei were stained with 4',6-diamidino-2-phenylindole (DAPI). Scale bar=40 μm. (c) Invasion assay of control (SCR) and TP53-silenced (p53−) NCM460 and HepG2 cells. The cells were exposed, 48 h after the end of the silencing procedure, to IL-6 for 24 h. (d) Real-time–PCR evaluation of 45S rRNA and western blot analysis of p53 expression in NCM460 and HepG2 cells transfected with control sequences (SCR) and in POLR1A-silenced cells (Pol1−). At 48 h after the end of the silencing procedure cells were exposed to IL-6 for 24 h. (e) Western blot and densitometric analysis of E-cadherin expression in control (SCR) and POLR1A-silenced (Pol1−) HepG2 cells. At 48 h after the end of the silencing procedure the cells were exposed to IL-6 for 24 h. (f) Invasion assay of control (SCR) and POLR1A-silenced NCM460 and HepG2 cells. At 48 h after the end of the silencing procedure, the cells were exposed to IL-6 for 24 h. Histograms show the values (mean±s.d.) of three experiments. *P<0.05; **P<0.01; n.s., not significant.
Also, the immunofluorescence visualization of SLUG and E-cadherin in NCM460 cells showed that IL-6 treatment increased SLUG expression, whereas it greatly reduced the E-cadherin-related signal (Figure 3b). All these data indicate that, in NCM460 and HepG2 cell lines, IL-6 induces phenotypic changes that are typical of EMT.27 We also investigated whether IL-6 might increase the cell invasiveness potential, and whether this was p53 dependent. For this purpose, we used control and TP53-silenced NCM460 and HepG2 cells (see the efficacy of TP53 silencing in Supplementary Figure 5). We seeded the cells in an invasion chamber and evaluated the effect of IL-6 treatment on their migration through the matrix for 24 h. We found that IL-6 enhanced the invasiveness of control cells without affecting that of TP53-silenced cells (Figure 3c and Supplementary Figure 6). Interestingly, the higher invasiveness observed in TP53-silenced cells in comparison with IL-6-stimulated cells was found to be associated with a lower level of E-cadherin expression in these cells (Supplementary Figure 7).
In order to rule out the possibility that IL-6 might activate other pathways leading to EMT, independent of rRNA synthesis stimulation, we investigated whether IL-6 was capable of inducing EMT in cells in which the interleukin could not enhance rRNA transcription. To avoid IL-6 stimulation of rRNA synthesis, we silenced the POLR1A gene coding for the RNA polymerase I catalytic subunit by the small-interfering RNA procedure in NCM460 and HepG2 cells (Supplementary Figure 8). This procedure induced rRNA synthesis inhibition without reducing protein synthesis11 and did not modify TP53 mRNA transcription (Supplementary Figure 9). The reduction of POLR1A mRNA expression was accompanied by a strong inhibition of rRNA synthesis and by the stabilization of p53. IL-6 treatment neither increased the synthesis of rRNA nor reduced the expression of p53 in POLR1A-silenced cells (Figure 3d). We then looked for changes in the E-cadherin expression in POLR1A-silenced HepG2 cells after IL-6 exposure: we observed no reduction in the expression of E-cadherin in comparison with unstimulated cells in POLR1A-silenced HepG2 cells (Figure 3e).
Furthermore, the invasion assay showed that IL-6 treatment did not increase the invasiveness of POLR1A-silenced cells (Figure 3f and Supplementary Figure 10). These results led us to conclude that the enhancement of rRNA synthesis was necessary for IL-6 to induce cells to undergo p53-mediated activation of the EMT program, whereas no other pathways that downregulate p53 expression and induce invasive neoplastic phenotypes were activated.
Changes in ribosome biogenesis, p53 and E-cadherin expression in the human epithelial cells of the colon with UC
The data obtained from experiments conducted using human epithelial cell lines indicated that the enhancement of rRNA synthesis by IL-6 exposure caused a downregulation of p53 expression with the consequent activation of the EMT program. We wondered whether similar changes occurred in the epithelial colon cells of patients with UC. We carried out this study on histological sections from tissue samples prepared for diagnostic purposes. In fact, for ethical reasons, we did not have the possibility of using fresh tissue samples. The results reported below are common to all the control and UC colon samples.
In order to evaluate whether an upregulation of rRNA transcriptional activity occurred in the epithelial cells of colonic mucosa from patients with UC, we measured the nucleolar size of these cells in histological sections selectively stained by silver for the argyrophilic nucleolar organizer region (AgNOR) proteins. This technique makes it possible to precisely evaluate the nucleolar size.28 As the nucleolar size is closely and directly related to rRNA transcription activity,15 we were able to obtain information on whether UC resulted in changes in the ribosome biogenesis rate. After silver staining, the histological characteristics of the normal colonic mucosa are perfectly recognized (Figure 4a). Colonic mucosa with UC appears to be characterized by a diffuse, mainly mononuclear inflammatory infiltrate in the lamina propria (Figure 4b). At a higher magnification, it is possible to observe that the epithelial cells of normal crypts exhibit small nucleoli (Figure 4c), whereas in crypt cells of UC they exhibited much larger nucleoli (Figure 4d). Morphometric analysis and statistical evaluation on normal and UC samples confirmed that in inflamed mucosa the size of crypt cell nucleoli was much larger than that of normal cell nucleoli. Hypertrophic nucleoli also characterized the surface epithelial cells of UC mucosa when compared with those of normal mucosa (Figures 4e and f). After treatment with anti-inflammatory drugs, the nucleolar size was reduced to the values of the epithelium of normal, control colonic mucosa (Figure 4g).
Ribosome biogenesis is upregulated in the epithelial cells of colon mucosa in UC, and returns to normal level after anti-inflammatory therapy. (a–g) Sections from histological routinely processed samples of human colon after the silver staining procedure for the selective visualization of nucleoli. At low magnification, the architecture of the normal (a) and UC mucosa (b) is perfectly recognizable. Note the higher cellularity in the lamina propria of the mucosa with UC, in comparison with the normal mucosa; *indicates lamina propria; scale bar=100 μm. At higher magnification, the nucleoli appeared to be darkly stained. The size of nucleoli of the epithelial cells lining the crypts of the normal mucosa (c) is smaller (mean nucleolar size=2.33±1.05 μm2) than that of the epithelial cell nucleoli in UC (mean nucleolar size=4.55±1.77 μm2) (d), P<0.0001. The same is true for the epithelial cells of the mucosal surface (1.27±0.44 μm2 vs 5.19±2.27 μm2, P<0.0001) (cross refer (e) with (f)). Scale bar=10 μm. In patients with UC treated with anti-inflammatory drugs (g) the hypercellularity was no longer present in the lamina propria (Scale bar=70 μm). Insets: the nucleoli of the epithelial cells lining the crypts and the surface of the mucosa exhibited the same size as that of the nucleoli in the normal mucosa, crypt cell nucleoli exhibiting a size of 2.45±0.99 μm2 and the surface epithelial cell nucleoli a size of 1.22±0.65 μm2. Scale bar=10 μm.
We then evaluated the expression of p53 in histological sections from control and inflamed colon mucosa biopsy samples. We found that in normal colon mucosa the labeling was present mainly in the epithelial cells of the deeper portion of the crypts (Figures 5a and b). The labeling index of epithelial cells within the crypts was 24.5 (±6.8 s.d.). Also, in the colon mucosa of patients with UC, labeled cells were mainly located in the deeper portion of the crypts (Figures 5c and d). However, the percentage of labeled cells was lower (labeling index=9.8±4.5 s.d.), thus indicating a downregulation of p53 expression in UC epithelial cells (P<0.01). These data are consistent with the recent observation reported by Risques et al.29 that p53 expression in human normal colon epithelial cells was higher, even though in a nonstatistically significant manner, than that of patients with UC in whom cancerous lesions had developed. After treatment with anti-inflammatory drugs, the percentage of labeled cells was found to be quite similar to that of normal, control colonic mucosa (27.2±7.9 s.d.) (Figures 5e and f). These changes were associated with quantitative variation of IL-6 expression in the inflammatory infiltrate. As shown in Supplementary Figure 11, the number of inflammatory cells immunostained for IL-6 was low in normal colon mucosa; it was increased in UC, and reverted to the normal colon value after treatment with anti-inflammatory drugs.
The p53 expression is downregulated in the epithelial cells of the colon mucosa in UC, and returns to normal level after anti-inflammatory therapy. (a–f) Histological sections from human colon treated with monoclonal anti-p53 antibodies visualized by the peroxidase/DAB enzymatic reaction. (a) Normal colon mucosa. A large number of epithelial cell nuclei in the deeper half portion of the crypts was brick-red stained, indicating a positive reaction for p53 accumulation. (b) Detail of (a), at higher magnification. (c) UC. Only a few epithelial cells appeared to be positively stained for p53. (d) Detail of (c), at higher magnification. (e) UC after anti-inflammatory therapy. The number of epithelial cell nuclei showing a positive p53 reaction was comparable to that of the epithelium of normal, control colonic mucosa. (f) Detail of (e), at higher magnification. Scale bar=100 μm (a, c and e). Scale bar=25 μm (b, d and f). Hematoxylin counterstaining.
We then visualized and analyzed the expression of E-cadherin in histological sections using the immunoperoxidase and immunofluorescence techniques. The analysis of E-cadherin expression using the immunoperoxidase technique indicated that the control mucosa samples were characterized by an intense signal, perfectly decorating the boundaries of all the epithelial cells in the crypts and on the surface (Figures 6a and c). Regarding UC samples, a focal reduction of the signal was observed (Figure 6b). Furthermore, in many crypts, whereas some clusters of epithelial cells appeared to exhibit E-cadherin staining, the contiguous epithelial cells were completely unstained (Figure 6d). In colon samples from patients treated with anti-inflammatory drugs, E-cadherin expression pattern was superimposable to that of normal, control colonic mucosa (Figure 6e). The same results were obtained with the immunofluorescence analysis (Supplementary Figures 12a–e).
E-cadherin staining is reduced in the epithelial cells of colon mucosa in UC, and returns to normal level after anti-inflammatory therapy. Immunostaining with anti-E-cadherin antibodies, revealed by a peroxidase/DAB enzymatic reaction. In normal mucosa, all the epithelial cells of the crypts and the surface exhibited an intense staining at their boundaries (scale bars=80 μm (a), 20 μm (c)). In UC mucosa, the staining intensity appeared to be strongly reduced in some zones (scale bars=80 μm (b), 20 μm (d)). Arrows indicate epithelial cells with low or nil E-cadherin staining. After anti-inflammatory therapy E-cadherin immunostaining was superimposable to that of normal colon mucosa (scale bar=80 μm (e)). Hematoxylin counterstaining.
Discussion
The present results highlight a new mechanism that may link chronic inflammation to cancer, based on p53 downregulation, which is activated by the enhancement of rRNA transcription upon IL-6 exposure. By using human cell lines we demonstrated that IL-6-stimulated c-MYC mRNA IRES translation. The consequent increase in c-myc protein expression was responsible for the upregulation of rRNA transcription. The enhanced rRNA transcription stimulated the MDM2-mediated degradation of p53 throughout the ribosome protein–MDM2–p53 pathway.12, 13 The reduction in p53 protein level was associated with a decrease in the function of p53, which activated the EMT program and increased the invasiveness potential of IL-6-exposed cells. The downregulation of p53 protein expression was exclusively due to the upregulated ribosome biogenesis. This was demonstrated by the observation that c-MYC silencing counteracted the stimulatory effect of IL-6 on rRNA transcription, whereas IL-6 exposure did not reduce p53 protein level in cells in which the rRNA transcription could not be stimulated as a consequence of RNA polymerase I-depletion. These effects appeared to be very likely independent of STAT3 activation. There is evidence that STAT3 has a crucial role in inflammation-associated tumorigenesis, namely in the colon and liver,5, 6, 7, 8 and IL-6 is the major activator of STAT3, which stimulates the transcription of target genes whose products increase cell proliferation and survival.30 On the other hand, our results suggest that the IL-6 effects reported in the present study were independent of the activation of the STAT3 pathway. In fact, consistently with the data reported on myeloma cell lines,17 we observed that IL-6 induced an enhanced expression of c-myc protein as early as 1 h after exposure by stimulating c-MYC mRNA IRES translation without modifying c-MYC mRNA transcription. Therefore, this ruled out the possibility that the increased level of c-myc protein might be the consequence of transcriptional changes of c-MYC mRNA expression due to STAT3 activation by IL-6. The increased c-MYC mRNA IRES translation was likely the effect of the IL-6 activation of the MAPK pathway.30 Indeed, in myeloma cell lines, IL-6 stimulation of the c-MYC-IRES function was induced by phosphorylation and the cytoplasmic localization of the RNA-binding protein heterogeneous nuclear ribonucleoproteins A1, an IRES-transactivating factor, in a MAPK-dependent fashion.31
The downregulation of p53 expression may be of primary importance in the process of cell transformation in inflamed tissues. In fact, together with the production of cytokines, chemokines and growth factors, which stimulate the proliferation and antiapoptotic activity, inflammatory cells generate reactive oxygen and nitrogen species,1 which cause DNA oxidative damages that may no longer be adequately repaired, because of the downregulated function of p53. This may lead to epigenetic alterations of both oncogenes and other tumor suppressor genes responsible for neoplastic transformation and progression. This mechanism may be operative in chronically inflamed tissues, such as the human colon mucosa with UC. Indeed, our histochemical and immunohistochemical findings obtained using colon biopsy samples from UC patients were quite consistent with the results from the study conducted on human cell lines.
In fact, we found that the epithelial cells of patients with UC disease were characterized by a nucleolar hypertrophy, which indicates an upregulated rRNA synthesis,15 a downregulation of p53 expression and a reduction of E-cadherin expression, which is a major phenotypic change indicating the activation of the EMT program.27 The IL-6-induced p53 downregulation mechanism of tumorigenesis in UC is shown schematically in Figure 7, as it may be suggested by the present findings. This mechanism is likely to be functionally active also in other inflammatory foci in which IL-6 is produced and in which IL-6 induces an upregulation of the nucleolar activity, similar to what happens in the epithelial cells of the surface and crypts of human colonic mucosa with UC. Indeed, there is evidence that human hepatocytes in hepatitis virus-related cirrhosis are characterized by a marked nucleolar hypertrophy, and it is worth noting that the number of hepatocytes with nucleolar hypertrophy was directly associated with an increased risk for hepatocellular carcinoma development.32
Suggested pathway linking chronic inflammation to cancer. In inflamed tissues, such as colon mucosa with UC, a high amount of IL-6 is produced. IL-6 upregulates c-myc protein expression, which, in turn, enhances ribosome biogenesis. The enhanced ribosome biogenesis is responsible for an increased MDM2-mediated p53 degradation. This, on one hand, may favor the EMT of the epithelial cells, and, on the other hand, may reduce the cell response to genotoxic DNA damages. Both consequences can greatly favor cancer onset.
Materials and methods
Patients
Samples of 10 patients (5 males, mean age 49.8±10.3 years) with UC and 5 healthy subjects (3 males, mean age 45.6±9.6 years) were used for the study. After informed consent, biopsy specimens were obtained during colonoscopy performed for a routine clinical care and surveillance. The diagnosis of UC was based on conventional clinical, endoscopic and pathohistological criteria as described by Lennard-Jones.33 Biopsies from patients without macroscopic abnormalities or histories of any gastrointestinal disease were used as the control group. All UC patients had suffered from pancolitis for more than 10 years. In all UC cases, specimens were obtained from the rectum and the descendent, transverse and ascending colon. All UC patients underwent an initial colonoscopy during active disease, and then a second endoscopy was performed during a clinical remission confirmed by histology. The remission of UC was obtained with corticosteroid therapy and afterwards maintained with mesalazine. The collected samples were fixed in formalin and embedded in paraffin for histological examination.
Cell lines and chemical treatments
HepG2, SW1990 and LS174T cell lines were obtained from American Type Culture Collection, and the NCM460 cell line was purchased from INCELL Corporation (San Antonio, TX, USA). All the cell lines were p53 wild-type. The HCT116 p53−/− cell line was a generous gift from Professor Bert Vogelstein.
The recombinant human IL-6 (Sigma-Aldrich, Milan, Italy) was used at a final concentration of 50 ng/ml; the proteasome inhibitor MG-132 (Calbiochem, Merck, Nottingham, UK) was used at a final concentration of 10 mM; cycloheximide (Sigma-Aldrich) was used at a concentration of 20 μg/ml; hydroxyurea (Sigma-Aldrich) was used at 3.4 mM; 5-FU (Fluorouracile, Teva Pharma Italia, Milan, Italy) was used at a dose of 20 μg/ml; Nutlin-3 (Sigma-Aldrich) was used at a concentration of 5 mM.
NOR silver staining
AgNOR staining and morphometric analysis were performed as described.15 For each case, the AgNOR area of at least 200 nuclei was measured and the mean (s.d.) AgNOR area was calculated.
Immunohistochemical assessment
A non-biotin-amplified method (NovoLinkTM Polymer Detection System, Novocastra Laboratories, Newcastle Upon Tyne, UK) was used for visualizing antigens in tissue sections. The sections were subsequently incubated with primary anti-E-cadherin (clone 32A8, Cell Signaling Technology, Beverly, MA, USA), anti-IL-6 (Sigma-Aldrich) and anti-p53 (Novocastra) antibodies diluted in 1% bovine serum albumin in Tris buffer saline overnight at 4 °C using the appropriate dilutions. The slices were then counterstained with hematoxylin, dehydrated and coverslipped. Nuclear p53 immunostaining was assessed by image cytometry, using the Cytometrica program (C&V, Bologna, Italy). Staining was expressed as the percentage of labeled nuclear area over the total nuclear area of epithelial cells in the section labeling index according to Faccioli et al.34
Immunofluorescence
Cells grown on glass coverslips were fixed and permeabilized in phosphate-buffered saline (PBS) containing 2% paraformaldehyde and 1% Triton X-100, while sections were dewaxed, hydrated through decreasing concentrations of ethanol, rinsed in distilled water, and subjected to antigen retrieval treatment. Samples were washed in PBS and then incubated in PBS and 1% bovine serum albumin (Sigma-Aldrich) to block nonspecific binding, before incubating with the primary anti-SLUG (clone L40C6, Cell Signaling Technology) and anti-E-cadherin (clone 32A8, Cell Signaling Technology) antibodies diluted in PBS and 1% bovine serum albumin overnight at 4 °C. Samples were subsequently rinsed in PBS and then incubated with fluorescein isothiocyanate (FITC)-conjugated anti-mouse secondary antibody (Dako). Mounting and nuclei counterstaining were performed using the ‘pro long antifade reagent with DAPI’ (Invitrogen, Carlsbad, CA, USA) and observed under a fluorescence microscope (Carl Zeiss, Milan, Italy).
Analysis of rRNA synthesis by 5-FU incorporation
Cells growing on coverslips were incubated for 15 min in medium containing 2 mM 5-FU (Sigma-Aldrich) and processed as described.11
RNA extraction, reverse transcription and real-time–PCR
Total RNA was extracted with TRI reagent (Ambion, Austin, TX, USA), and real-time–PCR measurements were performed as previously described.11 The relative amounts of TP53, 45S rRNA, BAX, PUMA, c-MYC, RPL11, POLR1A and β-glucuronidase mRNAs were evaluated by real-time–PCR. TP53, POLR1A and c-MYC and the internal control β-glucuronidase mRNAs were quantified with TaQMan Gene Expression Assays primers and probe kits (Applied Biosystems, Foster City, CA, USA); primers for SYBR Green real-time–PCR analysis of human 45S rRNA, RPL11, BAX and PUMA were designed using the Roche online primers design tool. All sequences are available on request.
Genes silencing by RNAi transfection
Select stealth RNAi (Invitrogen) targeted against TP53, c-MYC and RPL11 and the catalytic subunit of RNA polymerase I (POLR1A) were used. Control silenced cells (scrambled) were transfected with equivalent amounts of Stealth RNAi negative control (Invitrogen). Cells were transfected with lipofectamine RNAiMAX (Invitrogen) in opti-MEM medium (Invitrogen), according to the manufacturer’s procedures.
Immunoblotting and immunoprecipitation
The immunoblotting, isolation of nuclear protein fractions and immunoprecipitation procedures were performed as previously described.11 Antibodies used in this work were as follows: anti-p53 (clone BP53-12, Novocastra), anti-β-actin (clone AC-74, Sigma-Aldrich), anti-MDM2 (clone SMP14 and clone H-221, Santa Cruz Biotechnology), anti-L11 (clone 3A4A7, Invitrogen), anti-SLUG (clone L40C6, Cell Signaling Technology), anti-E-cadherin (clone 32A8, Cell Signaling Technology), anti-c-Myc (Cell Signaling Technology) and anti-Lamin B (C-20, Santa Cruz Biotechnology).
mRNA transfection
For RNA transfection, capped mRNA was transcribed from linearized pR-c-MYC-IRES-F (gift from Professor RJ Schneider) and a control pRF plasmid, as previously described,35 using the mMessage mMachine T3 kit (Ambion). Cells were transfected with 0.4 μg RNA/sample using Lipofectamine 2000 (Invitrogen) following the manufacturer’s instructions. After 8 h transfection, cells were harvested and analyzed with a dual-luciferase assay kit (Promega) according to the manufacturer’s instructions.
Cell invasion assay
Invasion assay was performed using Boyden chambers (New Technologies Group, Milan, Italy) containing a polyvinyl pyrrolidone-free polycarbonate filter with 8 μm pores. Membranes were coated with 15 μg Matrigel (Sigma-Aldrich). Cells were seeded in the upper chamber in serum-free medium in the presence or absence of IL-6 (50 ng/ml) for 24 h at 37 °C. Complete medium was placed in the lower compartment as a chemoattractant. After incubation, invading cells in the lower surface were fixed in ice-cold methanol, stained with toluidine blue staining (Sigma-Aldrich), and scored as the mean number of invaded cells per 10 random optical fields, in three independent experiments, at × 20 magnification.
Statistical analysis
The χ2 or Mann–Whitney U-test, when appropriate, was used for the comparisons among groups. Agreement between scores was assessed by κ statistics. All statistics were obtained using the SPSS statistical software package (SPSS, Inc.). P-values <0.05 were regarded as statistically significant.
References
Coussens LM, Werb Z . Inflammation and cancer. Historical key review on inflammation and cancer. Nature 2002; 420: 860–867.
Mantovani A, Allavena P, Sica A, Balkwill F . Cancer-related inflammation. Nature 2008; 454: 436–444.
Lin WW, Karin M . A cytokine-mediated link between innate immunity, inflammation, and cancer. J Clin Invest 2007; 117: 1175–1183.
Grivennikov SI, Greten FR, Karin M . Immunity, inflammation and cancer. Cell 2010; 140: 883–899.
Bollrath J, Phesse TJ, von Burstin VA, Putoczki T, Bennecke M, Bateman T et al. gp130-mediated Stat3 activation in enterocytes regulates cell survival and cell-cycle progression during colitis-associated tumorigenesis. Cancer Cell 2009; 15: 91–102.
Grivennikov S, Karin E, Terzic J, Mucida D, Yu GY, Vallabhapurapu S et al. IL-6 and Stat3 are required for survival of intestinal epithelial cells and development of colitis associated cancer. Cancer Cell 2009; 15: 103–113.
Park EJ, Lee JH, Yu GY, He G, Ali SR, Holzer RG et al. Dietary and genetic obesity promote liver inflammation and tumorigenesis by enhancing IL-6 and TNF expression. Cell 2010; 140: 197–208.
He G, Karin M . NF-κB and STAT3 - key players in liver inflammation and cancer. Cell Res 2011; 21: 159–168.
Kim S, Keku TO, Martin C, Galanko J, Woosley JT, Schroeder JC et al. Circulating levels of inflammatory cytokines and risk of colorectal adenomas. Cancer Res 2008; 68: 323–328.
Nakagawa H, Maeda S, Yoshida H, Tateishi R, Masuzaki R, Ohki T et al. Serum IL-6 levels and the risk for hepatocarcinogenesis in chronic hepatitis C patients: an analysis based on gender differences. Int J Cancer 2009; 125: 2264–2269.
Donati G, Bertoni S, Brighenti E, Vici M, Treré D, Volarevic S et al. The balance between rRNA and ribosomal protein synthesis up and downregulates the tumour suppressor p53 in mammalian cells. Oncogene 2011; 30: 3274–3288.
Zhang Y, Lu H . Signaling to p53: ribosomal proteins find their way. Cancer Cell 2009; 16: 369–377.
Deisenroth C, Zhang Y . Ribosome biogenesis surveillance: probing the ribosomal protein-Mdm2-p53 pathway. Oncogene 2010; 29: 4253–4260.
Terzić J, Grivennikov S, Karin E, Karin M . Inflammation and colon cancer. Gastroenterology 2010; 138: 2101–2114.
Derenzini M, Trere D, Pession A, Montanaro L, Sirri V, Ochs RL . Nucleolar function and size in cancer cells. Am J Pathol 1998; 152: 1291–1297.
Kiuchi N, Nakajima K, Ichiba M, Fukada T, Narimatsu M, Mizuno K et al. STAT3 is required for the gp130-mediated full activation of the c-myc gene. J Exp Med 1999; 189: 63–73.
Shi Y, Frost PJ, Hoang BQ, Benavides A, Sharma S, Gera JF et al. IL-6-induced stimulation of c-myc translation in multiple myeloma cells is mediated by myc internal ribosome entry site function and the RNA-binding protein, hnRNP A1. Cancer Res 2008; 68: 10215–10222.
Zhong Z, Wen Z, Darnell JE Jr . Stat3: a STAT family member activated by tyrosine phosphorylation in response to epidermal growth factor and interleukin-6. Science 1994; 264: 95–98.
Niu G, Wright KL, Ma Y, Wright GM, Huang M, Irby R et al. Role of Stat3 in regulating p53 expression and function. Mol Cell Biol 2005; 25: 7432–7440.
Vassilev LT, Vu BT, Graves B, Carvajal D, Podlaski F, Filipovic Z et al. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science 2004; 303: 844–848.
Sax JK, El-Deiry WS . p53 downstream targets and chemosensitivity. Cell Death Differ 2003; 10: 413–417.
Moran DM, Mattocks MA, Cahill PA, Koniaris LG, McKillop IH . Interleukin-6 mediates G(0)/G(1) growth arrest in hepatocellular carcinoma through a STAT3-dependent pathway. J Surg Res 2008; 147: 23–33.
Christofori G, Semb H . The role of the cell-adhesion molecule E-cadherin as a tumour-suppressor gene. Trends Biochem Sci 1999; 24: 73–76.
Perl AK, Wilgenbus P, Dahl U, Semb H, Christofori G . A causal role for E-cadherin in the transition from adenoma to carcinoma. Nature 1998; 392: 190–193.
Bolós V, Peinado H, Pérez-Moreno MA, Fraga MF, Esteller M, Cano A . The transcription factor Slug represses E-cadherin expression and induces epithelial to mesenchymal transitions: a comparison with Snail and E47 repressors. J Cell Sci 2003; 116: 499–511.
Wang SP, Wang WL, Chang YL, Wu CT, Chao YC, Kao SH et al. p53 controls cancer cell invasion by inducing the MDM2-mediated degradation of Slug. Nat Cell Biol 2009; 11: 694–704.
Polyak K, Weinberg RA . Transitions between epithelial and mesenchymal states: acquisition of malignant and stem cell traits. Nat Rev Cancer 2009; 9: 265–273.
Montanaro L, Treré D, Derenzini M . Nucleolus, ribosomes, and cancer. Am J Pathol 2008; 173: 301–310.
Risques RA, Lai LA, Himmetoglu C, Ebaee A, Li L, Feng Z et al. Ulcerative colitis-associated colorectal cancer arises in a field of short telomeres, senescence, and inflammation. Cancer Res 2011; 7: 1669–1679.
Yu H, Pardoll D, Jove R . STATs in cancer inflammation and immunity: a leading role for STAT3. Nat Rev Cancer 2009; 9: 798–809.
Shi Y, Frost P, Hoang B, Benavides A, Gera J, Lichtenstein A . IL-6-induced enhancement of c-Myc translation in multiple myeloma cells: critical role of cytoplasmic localization of the rna-binding protein hnRNP A1. J Biol Chem 2011; 286: 67–78.
Trerè D, Borzio M, Morabito A, Borzio F, Roncalli M, Derenzini M . Nucleolar hypertrophy correlates with hepatocellular carcinoma development in cirrhosis due to HBV infection. Hepatology 2003; 37: 72–78.
Lennard-Jones JE . Classification of inflammatory bowel disease. Scand J Gastroenterol Suppl 1989; 170: 2–6.
Faccioli S, Chieco P, Gramantieri L, Stecca BA, Bolondi L . Cytometric measurement of cell proliferation in echo-guided biopsies from focal lesions of the liver. Mod Pathol 1996; 9: 120–125.
Rocchi L, Pacilli A, Sethi R, Penzo M, Schneider RJ, Treré D et al. Dyskerin depletion increases VEGF mRNA internal ribosome entry site-mediated translation. Nucleic Acids Res 2013; 41: 8308–8318.
Acknowledgements
This work was supported by the Roberto and Cornelia Pallotti’s Legacy for Cancer Research, the Vanini–Cavagnino Grant (Centro Interdipartimentale per le Ricerche sul Cancro, Bologna, Italy), the Italian Ministry of Education, University and Research (MIUR: grants for orientated fundamental research) and Associazione Italiana per la Ricerca sul Cancro (AIRC, grant n° IG13480).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Oncogene website
Supplementary information
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/3.0/
About this article
Cite this article
Brighenti, E., Calabrese, C., Liguori, G. et al. Interleukin 6 downregulates p53 expression and activity by stimulating ribosome biogenesis: a new pathway connecting inflammation to cancer. Oncogene 33, 4396–4406 (2014). https://doi.org/10.1038/onc.2014.1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/onc.2014.1
Keywords
This article is cited by
-
Cancer-associated fibroblasts induce growth and radioresistance of breast cancer cells through paracrine IL-6
Cell Death Discovery (2023)
-
Tumor suppressor p53 mediates interleukin-6 expression to enable cancer cell evasion of genotoxic stress
Cell Death Discovery (2023)
-
Role of the E3 ubiquitin-ligase Hakai in intestinal inflammation and cancer bowel disease
Scientific Reports (2022)
-
Green tea EGCG effectively alleviates experimental colitis in middle-aged male mice by attenuating multiple aspects of oxi-inflammatory stress and cell cycle deregulation
Biogerontology (2022)
-
AMG-232 sensitizes high MDM2-expressing tumor cells to T-cell-mediated killing
Cell Death Discovery (2020)