Abstract
The colony-stimulating factor-1 (CSF-1) and its receptor CSF-1R physiologically regulate the monocyte/macrophage system, trophoblast implantation and breast development. An abnormal CSF-1R expression has been documented in several human epithelial tumors, including breast carcinomas. We recently demonstrated that CSF-1/CSF-1R signaling drives proliferation of breast cancer cells via ‘classical’ receptor tyrosine kinase signaling, including activation of the extracellular signal-regulated kinase 1/2. In this paper, we show that CSF-1R can also localize within the nucleus of breast cancer cells, either cell lines or tissue specimens, irrespectively of their intrinsic molecular subtype. We found that the majority of nuclear CSF-1R is located in the chromatin-bound subcellular compartment. Chromatin immunoprecipitation revealed that CSF-1R, once in the nucleus, binds to the promoters of the proliferation-related genes CCND1, c-JUN and c-MYC. CSF-1R also binds the promoter of its ligand CSF-1 and positively regulates CSF-1 expression. The existence of such a receptor/ligand regulatory loop is a novel aspect of CSF-1R signaling. Moreover, our results provided the first evidence of a novel localization site of CSF-1R in breast cancer cells, suggesting that CSF-1R could act as a transcriptional regulator on proliferation-related genes.
Similar content being viewed by others
Introduction
The colony-stimulating factor-1 (CSF-1) was first described as a hematopoietic growth factor regulating key functions of monocytes and macrophages through the activation of the class III receptor tyrosine kinase (RTK) CSF-1R.1, 2 It is well established that the CSF-1/CSF-1R pathway has regulatory roles also outside the hematopoietic system.3, 4, 5 The abnormal expression of CSF-1R, with or without that of CSF-1, has been reported in several cancers and cancer-derived cell lines.3, 6, 7 In particular, CSF-1/CSF-1R overexpression is associated with poor prognosis8, 9 and is predictive of ipsilateral recurrence in breast cancer patients.10
CSF-1-induced activation of CSF-1R promotes receptor dimerization and tyrosine transphosphorylation in the intracellular kinase domain, resulting in the activation of downstream signaling pathways.11 In addition to this ‘classical’ RTK signaling, increasing evidences support the translocation of RTK from the plasma membrane to the nucleus.12 In particular, epidermal growth factor receptor family members,13, 14, 15 fibroblast growth factor receptors 1 and 3,16, 17, 18 insulin and insulin-like growth factor-1 receptors19, 20 and the vascular endothelial growth factor receptor 2,21, 22 localize within the nucleus, as either full-length receptors or cleaved fragments, with or without their ligands. Once in the nucleus, RTK can regulate the expression of target genes, such as CCND1,13, 23 FGF2,24 COX215 and c-Jun.23
We, along with others, have previously showed that CSF-1R is expressed in breast cancer cell lines and tissues.7, 25, 26 Importantly, we demonstrated that CSF-1/CSF-1R signaling can drive cell proliferation of breast cancer cells via the activation of extracellular signal-regulated kinase 1/2 and the subsequent regulation of c-Jun, cyclin D1 and c-Myc expression.7 Here we demonstrated that in breast cancer cells (i) CSF-1R localizes in the nucleus, (ii) nuclear CSF-1R binds to the promoter region of proliferation-related genes and (iii) CSF-1R regulates the transcription of its ligand CSF-1.
Results and discussion
CSF-1R localizes in the nucleus of breast cancer cells
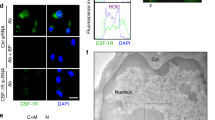
We recently reported that CSF-1R mRNA is expressed in breast cancer cells and this is matched by protein expression at the cell surface.7 Among the breast cancer cell lines analyzed, SKBR3 cells express the highest levels of membrane-bound CSF-1R and produce CSF-1 that sustains an autocrine proliferative loop.7 While assessing CSF-1R expression in SKBR3 cells by western blotting, we found that the composition of lysis buffer dramatically affected CSF-1R protein yield (Figure 1a). In particular, Laemmli buffer allowed the recovery of the highest amount of CSF-1R, while either radioimmunoprecipitation assay buffer or Frackelton buffers resulted in incomplete CSF-1R solubilization. In this respect, it should be noted that Frackelton buffer, which contains the non-ionic Triton X-100 as the only detergent, mainly extracts integral plasma membrane proteins and is normally used for CSF-1R signaling studies. By contrast, the type of buffer did not significantly affect CSF-1R recovery from lysates of RAW264.7 or BAC1.2F5 murine macrophages that express high levels of CSF-1R, or NIH/3T3 fibroblasts stably transfected (NIH/3T3-Fms cells) in order to express human CSF-1R (Figure 1a). These results suggested that CSF-1R solubility in SKBR3 breast cancer cells is different from that in macrophages or fibroblasts ectopically expressing CSF-1R.
CSF-1R localizes in the nucleus of SKBR3 breast cancer cells. (a) Effects of the composition of lysis buffer on CSF-1R protein yield. BAC1.2F5 and RAW264.7 (RAW264) murine macrophages,41 NIH/3T3 murine fibroblasts expressing or not ectopic human Fms (NIH/3T3-Fms; kind gift of MF Roussel, St. Jude Children’s Research Hospital, Memphis, TN, USA)42 and the breast cancer cell lines SKBR3 (HER2 positive), MDAMB468 (triple negative, basal-like 1; see below) and MDAMB231 (triple negative, mesenchymal stem like; see below)43, 44 were cultured in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum. L-cell conditioned medium was added (15%) to BAC1.2F5 cells as a source of CSF-1.45 Cells were lysed in Laemmli,7 complete radioimmunoprecipitation assay buffer (RIPA)46 or Frackelton47 lysis buffer. Proteins were subjected to immunoblotting using rabbit α-CSF-1R (C-20, raised against the C terminus of CSF-1R, Santa Cruz, Santa Cruz Biotechnology, Inc., Heidelberg, Germany, sc-692) or mouse α-vinculin (Sigma, Sigma-Aldrich, S.r.l. Milano, Italy, V9131) antibodies. (b) Determination of CSF-1R intracellular localization by immunofluorescence (IF). Routinely cultured cells were subjected to IF using a rabbit α-CSF-1R antibody48 and nuclei stained with the Hoechst dye. Scale bars: 20 μm. (c) CSF-1R localization in SKBR3 cells by cell fractionation. Gene silencing was performed with 50 nM SMART-pool small interfering RNA (siRNA) targeting CSF-1R mRNA (NM_005211 mRNA, Dharmacon, Thermo Fisher Scientific Inc., Rockford, IL, USA, number M-003109-03) or 50 nM siCONTROL non-targeting pool (siNT, Dharmacon, number D-001206-13) as previously described.48 Three days after transfection, cell fractionation was performed according to the manufacturer’s instructions using the Subcellular Protein Fractionation Kit for Cultured Cells (Pierce, Thermo Fisher Scientific Inc., Rockford, IL, USA, number 78840), which allows the subsequent separation of the cytosol, membrane, soluble and chromatin-bound nuclear fractions and cytoskeleton. Extracted proteins were subjected to immunoblotting with a rabbit α-CSF-1R antibody. Cell fractionation accuracy for membrane (ME), nuclear soluble (NE) and chromatin-bound (CBE) extracts was assessed by immunoblotting with α-fibrillarin (chromatin-bound marker; Santa Cruz, D-14, number sc-11336), α-HER2 (membrane marker; Cell Signaling, Technology, Danvers, MA, USA, number 2242), α-HSP90 (membrane/cytosol marker; Santa Cruz, number sc-13199) or α-HDAC2 (nuclear marker; Santa Cruz C-19, number sc-6296) antibodies. Immunoblotting with a marker for cytoskeleton (vimentin) did not produce any significant signal within nuclear compartments (not shown). (d) Effects of CSF-1R silencing in SKBR3 cells on nuclear CSF-1R. Gene silencing was performed as reported above using siGLO red (Cy3-labeled siGLO RISC-free siRNA, Dharmacon, number D-001600-01) as a transfection efficiency read-out following the manufacturer’s instructions. Three days after transfection, IF was performed with a rabbit α-CSF-1R antibody; fluorescence in the cytosol (cyto) or the nucleus (nuc) was quantified using ImageJ (graph) (http://rsb.info.nih.gov/ij/). Histograms represent means±s.e.m.; Student’s t-test: ***P<0.001 (n=2, 20 cells were quantified). Scale bars: 20 μm. Migration of molecular weight markers is indicated on the left.
To test whether differences of solubility were due to a different distribution of CSF-1R among cell compartments, we performed immunofluorescence in confocal microscopy, observing that CSF-1R localizes in the plasma membrane, cytoplasm and nucleus of SKBR3 cells (Figure 1b). In contrast, in keeping with a previous work,27 CSF-1R localized in the plasma membrane, cytosol and Golgi, but not in the nucleus of fibroblasts (NIH3T3-Fms) or macrophages (BAC1.2F5 and RAW264.7) (Figure 1b and Supplementary Figure 1).28 We then carried out subcellular fractionation of SKBR3 cells using a protocol that allows to discriminate between the soluble and chromatin-bound nuclear fractions as well as the plasma membrane (see figure legends for further details). The full-length mature form of CSF-1R was the prevailing protein recovered from the nucleus and in particular from the chromatin-bound nuclear fraction (Figure 1c, see also Supplementary Figure 2e). The cross-contamination between subcellular compartments was excluded using established controls.
A possible nuclear localization of CSF-1R was suggested by immunohistochemical studies of cervical pre-neoplastic tissues.29 Our paper is the first to report the localization of CSF-1R within the nucleus in the chromatin-bound compartment, a phenomenon occurring in breast cancer cells selectively (see also above). Nuclear localization of CSF-1R was indeed undetectable in other CSF-1R-expressing cells, such as macrophages, where, by contrast, CSF-1R localization at the nuclear envelope has been reported.30
CSF-1R silencing using small interfering RNA in SKBR3 cells (Figures 1c and d) reduced CSF-1R protein levels in the nuclear fractions, as assessed by western blotting or immunofluorescence. CSF-1R silencing efficiency shown in Figures 1c and d was at least 50%, as previously reported.7 The fact that the data obtained by western blotting were confirmed by immunofluorescence (Figure 1d) highlights the reliability of confocal imaging in studies of CSF-1R subcellular localization.
The mechanism of RTK trafficking to the nucleus has not been fully clarified yet. A mechanism involving cognate ligand binding and signaling proteins such as PI3K and Rab5 has been described31 and could explain CSF-1R translocation to the nuclear envelope.30 However, the steps required for CSF-1R to enter into the nucleus are still unknown. Nuclear localization signals (NLS) have been identified in RTK.32 To determine whether CSF-1R contains putative NLS, we used the online database NucPred (http://www.sbc.su.se/~maccallr/nucpred).33 NucPred assigns a score equal to 1 to an amino-acid sequence that is recognized as a bona fide NLS. When the sequence of c-erb3, which localizes within the nucleus,34 was run as a control, a score of 0.58 was obtained. When CSF-1R was run, no NLS was identified (score 0.15). By contrast, when CSF-1 (Gene ID: 1435; isoform 1) was run, the NLS score obtained (0.63) was similar to that found for c-erb3. In CSF-1, indeed, a cluster of three basic amino acids (arginine residues 521–524) showed NLS properties. This cluster was also present in CSF-1 isoforms 2 (438 amino acids) and 3 (256 amino acids), alternative splicing variants of the 554 amino acid-long proteoglycan precursor. No NLS was predicted (score 0.22) in interleukin-34, a recently described CSF-1R ligand.35 Previous studies indicated that CSF-1 is produced by breast cancer cell lines7 and may be located in the nucleus in breast cancer tissues.9 Thus, the high NLS score of CSF-1 led us to speculate that CSF-1R could translocate into the nucleus together with CSF-1. This hypothesis was strengthened by confocal immunofluorescence showing nuclear colocalization of CSF-1 and CSF-1R (Supplementary Figures 2a–c). Interestingly, biochemical fractionation revealed that an ∼32 kDa CSF-1 form, which may well correspond to membrane CSF-1, was the prevailing form in the cytosol/membrane fraction. In contrast, the ∼45 kDa form enriched in the nuclear fraction may represent different CSF-1 forms36 (Supplementary Figure 2d). Therefore, it is difficult to predict which form of CSF-1 is involved in CSF-1R nuclear trafficking. It should be noted that the cluster of arginine residues responsible for NLS is in the C-terminal domain of CSF-1 so that it would be relevant only in membrane-spanning CSF-1 precursor but not in the secreted mature forms.37 Nevertheless, CSF-1R nuclear translocation seems to be CSF-1 dependent, as nuclear CSF-1R increased following CSF-1 administration to serum-starved SKBR3 cells (Supplementary Figure 2e). The CSF-1-dependent, NLS-mediated nuclear translocation of CSF-1R may be only one of the mechanisms driving CSF-1R nuclear trafficking. For instance, Fms-interacting protein, which includes a NLS, binds transiently to the cytoplasmic domain of, and is phosphorylated on tyrosine by, activated CSF-1R.38 This may result in CSF-1R nuclear translocation. Further experiments are needed to address the mechanism of CSF-1R entry into the nucleus.
CSF-1R localizes in the nucleus of breast cancer cell lines and tissue irrespective of the intrinsic molecular subtypes
The nuclear localization of CSF-1R is not restricted to SKBR3 cells, as it was confirmed in other breast cancer cell lines, such as MDAMB231 and MDAMB468 (Figure 2a). Moreover, the colocalization of CSF-1R with nucleolin seems to indicate that, once in the nucleus, CSF-1R may localize also within the nucleolus (Figure 2a). Notably, the same subcellular distribution of CSF-1R was observed using two different antibodies raised against the C- or N terminus of CSF-1R (Supplementary Figure 3). These results are also in keeping with the presence of full-length CSF-1R within the nucleus (Figure 1c and Supplementary Figure 2c) and with what was observed for CSF-1R at the nuclear envelope.30 However, we sequenced CSF1R mRNA in SKBR3 cells in order to exclude nucleotide changes that could justify different properties of CSF-1R protein, including acquisition of NLS. CSF1R mRNA in SKBR3 cells, when compared with wild-type CSF1R sequence (ENSG00000182578.9), exhibited two single nucleotide changes (83A>G; 726G>A) that are conservative (Supplementary Figure 4). Next, the rabbit α-CSF-1R antibody was optimized for immunohistochemical staining (Supplementary Figure 1) and used to stain samples of invasive breast cancers. Figure 2b shows representative images of the nuclear and/or cytoplasmic immunohistochemical staining for CSF-1R in tissue specimens derived from breast cancer patients with different intrinsic molecular subtypes. We found that 40 out of 42 samples of breast cancers expressed CSF-1R; of these, 9 out of 40 expressed CSF-1R in both the nucleus and the cytosol, 3 out of 40 in the nucleus only and 28 out of 40 in the cytosol only (Supplementary Table 1). Notably, despite the limited number and heterogeneity of tissue specimens studied, we found that the nuclear expression of CSF-1R negatively correlates with progesterone receptor expression (rs=−0.37, P=0.018). The size of our cohort of patients could not provide the statistical power to establish the prognostic and/or predictive value of nuclear CSF-1R in breast cancer. However, as progesterone receptor expression correlates with a favorable prognosis in breast cancer, our data support previous reports where CSF-1R expression was related to poor prognosis.8, 9, 10
Nuclear localization of CSF-1R in breast cancer cell lines and tissue samples. (a) Immunofluorescence (IF) of CSF-1R in breast cancer cell lines. SKBR3, MDAMB231 and MDAMB468 cells were subjected to IF using rabbit α-CSF-1R and α-nucleolin (Santa Cruz, number sc-8031) antibodies. (b) Immunohistochemistry of CSF-1R in invasive breast cancers. Antigen retrieval was induced with sodium citrate buffer (pH 6.0) for 20 min at 97°C, rabbit α-CSF-1R antibody applied at 1:500 dilution and detection performed with labeled polymer (EnVision, DAKO Italia S.p.A., Milano, Italy). Examples of nuclear (arrowheads), cytosolic (dashed-line arrows) or cytosolic and nuclear (arrows) are indicated. Scale bars: 20 μm. Case (#) details are reported in Supplementary Table 1. The Research Ethics Committee of the Prato Hospital approved the use of breast cancer samples for this study (Protocol number 7741, 10 April 2012).
Feedback regulation of CSF-1/CSF-1R signaling in SKBR3 cells: CSF-1R regulates the transcription of CSF-1 and proliferation genes
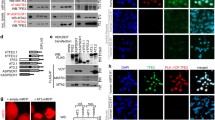
We previously showed that ‘classical’ RTK signaling exists in breast cancer cells where CSF-1R is activated by its ligand CSF-1, leading to CSF-1-induced proliferation and expression of genes involved in cell growth such as CCND1, c-MYC and c-JUN.7 Previous reports demonstrated that CCND1 and c-JUN promoters are targets of other nuclear RTK.31 Consequently, we investigated whether CSF-1R could bind the promoter of CCND1, c-JUN and c-MYC by chromatin immunoprecipitation. Chromatin immunoprecipitation was performed in SKBR3 cells using two different α-CSF-1R antibodies. As revealed by reverse transcriptase–PCR (Figure 3a) and quantified by quantitative PCR (Figures 3b and d), CSF-1R bound the promoter regions of CCND1, c-MYC and c-JUN. The reliability of chromatin immunoprecipitation procedure was supported by the absence of CSF-1R binding to GADPH promoter region (which is constitutively open and prone to transcription) or to an inaccessible region used as a negative control (Figures 3e and f). CSF-1R binding to gene promoters is a novel finding and may represent an additional mechanism of regulation of well-known targets downstream CSF-1R.39 Of note, the increased nuclear localization of epidermal growth factor receptor in tumors is associated with treatment resistance and poor prognosis.40
CSF-1R binds to the promoters of proliferation-related genes in SKBR3 cells. Chromatin immunoprecipitation (ChIP) was performed as previously described.49, 50, 51 Immunoprecipitation (IP) was performed with the indicated antibodies: rabbit α-C-terminal-CSF-1R (Ct-CSF-1R; 0.4 μg per sample), mouse α-N-terminal-CSF-1R (Nt-CSF-1R; B-8, Santa Cruz, number sc-46662, 4 μg per sample), control rabbit IgG (rIgG; 0.4 μg per sample, Sigma number G5518), control mouse IgG (mIgG; 4 μg per sample, number M7023). Input and a negative control for the IP procedure (that is, without antibody; C-(IP)) are shown. The primers used were as follows: CCND1prom fw 5′-GAGGGGACTAATATTTCCAGCAA-3′, rev 5′-TAAAGGGATTTCAGCTTAGCA-3′; c-Myc prom fw 5′-agggcttctcagaggcttg-3′, rev 5′-cctattcgctccggatctc-3′; c-JUN prom fw 5′-AAAGCTATGTATGTATGTGCTGCAT-3′, rev 5′-AACCGAGAGAACCTTCCTTTTTAT-3′; GAPDH prom fw 5′-TACTAGCGGTTTTACGGGCG-3′, rev 5′-TCGAACAGGAGGAGCAGAGAGCGA-3′; negative ChIP control fw 5′-ATGGTTGCCACTGGGGATCT-3′, rev 5′-TGCCAAAGCCTAGGGGAAGA-3′. Reverse transcriptase–PCR (a) and quantitative PCR (b–f) for the promoter regions of the indicated genes were performed as previously described.7, 51 Histograms represent the relative quantification of DNA recovered from IP with the indicated antibodies. Values were intra-experimentally normalized for input DNA and expressed as fold-change with respect to control IgG. Values are means±s.e.m. of data from three independent experiments. Student’s t-test comparing CSF-1R IP with the relative IgG: ***P<0.001.
The expression of CSF-1R together with CSF-1 has been reported in several cancers and cancer-derived cell lines.3, 6, 7 On the other hand, nuclear RTK may regulate the expression of cognate ligands by binding to their promoters.31 As revealed by reverse transcriptase–PCR (Figure 4a) and quantified by quantitative PCR (Figure 4b) in chromatin immunoprecipitation experiments, CSF-1R bound the promoter region of CSF1 in SKBR3 cells. CSF-1R silencing (Figure 4c) reduced significantly the amount of CSF-1R bound to the CSF1 promoter region (Figure 4d), supporting our finding. Furthermore, CSF-1R silencing resulted in a ‘more closed’ conformation of CSF1 promoter, as indicated by the reduction of acetylated H4 and RNA-polymerase-II bound to the CSF1 promoter region and the increase of bound DNA-methyltransferase-1 (Figure 4e). Accordingly, CSF1 expression decreased after CSF-1R silencing (Figure 4f). These data indicated that CSF-1R drives a ‘self-sustaining’ loop of CSF-1R signaling in breast cancer cells by regulating CSF-1 expression. Further experiments, however, need to address how CSF-1R work as a transcriptional regulator. In this respect, it should be noted that transcriptional activity has been found for other RTK.31
CSF-1R binds to the promoter of CSF1 and regulates its transcription in SKBR3 cells. Chromatin immunoprecipitation was performed (a, c) using the antibodies indicated in Figure 3 or rabbit α-pan-acetylated H4 (acH4; Millipore, S.p.A., Milano, Italy, number 06-598), mouse α-RNA-polymerase-II (Abcam, Cambridge, UK, number ab5408) or rabbit α-DNA-methyltransferase-1 (Abcam, number ab5208). CSF1 promoter region was amplified by PCR using the following primers: CSF1prom fw 5′-CACGAGGGAGCAAGTAACAC-3′; rev 5′-AGCCTTCAGCAAACGAG-3′. (a) Reverse transcriptase (RT)–PCR for CSF1 promoter region. INPUT and a negative control for the IP procedure (that is, without antibody; C-(IP)) are shown. (b, d, e) Quantitative (Q)-PCR for CSF1 promoter region. Histograms represent the relative quantification (RQ) of DNA recovered from IP with the indicated antibodies from routinely cultured cells (b) or 72 h after transfection with the indicated small interfering RNA (d, e). Values were normalized for input DNA in each experiment and expressed as fold-change with respect to control IgG. (c, f) Effects of CSF1R silencing on CSF1 expression. Q-PCR from DNA obtained 72 h after transfection with siNT (light gray) or siCSF1R (dark grey). RNA extraction, complementary DNA synthesis, quantitative and RT–PCR were performed as previously described.7, 49, 50, 51 The primers used were the following: CSF1 mRNA fw 5′-ATGACAGACAGGTGGAACTGCCAG-3′, rev 5′-TCACACAACTTCAGTAGGTTCAGG-3′; CSF1R mRNA (N-term) fw 5′-GGAGGCTGCCCAGATCGT-3′, rev 5′-GCGAGCTTGGTGTTGTTGTG-3′; and CSF1R mRNA (C-term) fw 5′-CCTCGCTTCCAAGAATTGCA-3′, rev 5′-CCCAATCTTGGCCACATGA-3′; histograms represent the RQ of mRNA using GAPDH or 18S to normalize data and siNT as calibrator. Values are means±s.e.m. of data from three independent experiments. Student’s t-test: *P<0.05; **P<0.01; ***P<0.001.
In conclusion, our data highlighted a novel aspect of CSF-1R function. Nuclear CSF-1R could work in parallel, and synergizes with, the classical RTK activity of CSF-1R. Further investigations have to be directed to determine whether nuclear CSF-1R is a druggable target and/or is suitable as a prognostic or predictive factor in breast cancer.
References
Sherr CJ, Rettenmier CW, Sacca R, Roussel MF, Look AT, Stanley ER . The c-fms proto-oncogene product is related to the receptor for the mononuclear phagocyte growth factor, CSF-1. Cell 1985; 41: 665–676.
Stanley ER, Chen DM, Lin HS . Induction of macrophage production and proliferation by a purified colony stimulating factor. Nature 1978; 274: 168–170.
Ide H, Seligson DB, Memarzadeh S, Xin L, Horvath S, Dubey P et al. Expression of colony-stimulating factor 1 receptor during prostate development and prostate cancer progression. Proc Natl Acad Sci USA 2002; 99: 14404–14409.
Kacinski BM . CSF-1 and its receptor in breast carcinomas and neoplasms of the female reproductive tract. Mol Reprod Dev 1997; 46: 71–74.
Toy EP, Azodi M, Folk NL, Zito CM, Zeiss CJ, Chambers SK . Enhanced ovarian cancer tumorigenesis and metastasis by the macrophage colony-stimulating factor. Neoplasia 2009; 11: 136–144.
Kirma N, Hammes LS, Liu YG, Nair HB, Valente PT, Kumar S et al. Elevated expression of the oncogene c-fms and its ligand, the macrophage colony-stimulating factor-1, in cervical cancer and the role of transforming growth factor-beta1 in inducing c-fms expression. Cancer Res 2007; 67: 1918–1926.
Morandi A, Barbetti V, Riverso M, DelloSbarba P, Rovida E . The colony-stimulating factor-1 (CSF-1) receptor sustains ERK1/2 activation and proliferation in breast cancer cell lines. PLoS One 2011; 6: e27450.
Kluger HM, Dolled-Filhart M, Rodov S, Kacinski BM, Camp RL, Rimm DL . Macrophage colony-stimulating factor-1 receptor expression is associated with poor outcome in breast cancer by large cohort tissue microarray analysis. Clin Cancer Res 2004; 10: 173–177.
Scholl SM, Pallud C, Beuvon F, Hacene K, Stanley ER, Rohrschneider L et al. Anti-colony-stimulating factor-1 antibody staining in primary breast adenocarcinomas correlates with marked inflammatory cell infiltrates and prognosis. J Natl Cancer Inst 1994; 86: 120–126.
Maher MG, Sapi E, Turner B, Gumbs A, Perrotta PL, Carter D et al. Prognostic significance of colony-stimulating factor receptor expression in ipsilateral breast cancer recurrence. Clin Cancer Res 1998; 4: 1851–1856.
Hamilton JA . CSF-1 signal transduction. J Leukoc Biol 1997; 62: 145–155.
Wang YN, Yamaguchi H, Hsu JM, Hung MC . Nuclear trafficking of the epidermal growth factor receptor family membrane proteins. Oncogene 2010; 29: 3997–4006.
Lin SY, Makino K, Xia W, Matin A, Wen Y, Kwong KY et al. Nuclear localization of EGF receptor and its potential new role as a transcription factor. Nat Cell Biol 2001; 3: 802–808.
Offterdinger M, Georget V, Girod A, Bastiaens PI . Imaging phosphorylation dynamics of the epidermal growth factor receptor. J Biol Chem 2004; 279: 36972–36981.
Wang SC, Lien HC, Xia W, Chen IF, Lo HW, Wang Z et al. Binding at and transactivation of the COX-2 promoter by nuclear tyrosine kinase receptor ErbB-2. Cancer Cell 2004; 6: 251–261.
Chioni AM, Grose R . FGFR1 cleavage and nuclear translocation regulates breast cancer cell behavior. J Cell Biol 2012; 197: 801–817.
Johnston CL, Cox HC, Gomm JJ, Coombes RC . Fibroblast growth factor receptors (FGFRs) localize in different cellular compartments. A splice variant of FGFR-3 localizes to the nucleus. J Biol Chem 1995; 270: 30643–30650.
Stachowiak MK, Maher PA, Joy A, Mordechai E, Stachowiak EK . Nuclear localization of functional FGF receptor 1 in human astrocytes suggests a novel mechanism for growth factor action. Brain Res Mol Brain Res 1996; 38: 161–165.
Sehat B, Tofigh A, Lin Y, Trocme E, Liljedahl U, Lagergren J et al. SUMOylation mediates the nuclear translocation and signaling of the IGF-1 receptor. Sci Signal 2010; 3: ra10.
Seol KC, Kim SJ . Nuclear matrix association of insulin receptor and IRS-1 by insulin in osteoblast-like UMR-106 cells. Biochem Biophys Res Commun 2003; 306: 898–904.
Feng Y, Venema VJ, Venema RC, Tsai N, Caldwell RB . VEGF induces nuclear translocation of Flk-1/KDR, endothelial nitric oxide synthase, and caveolin-1 in vascular endothelial cells. Biochem Biophys Res Commun 1999; 256: 192–197.
Mayo LD, Kessler KM, Pincheira R, Warren RS, Donner DB . Vascular endothelial cell growth factor activates CRE-binding protein by signaling through the KDR receptor tyrosine kinase. J Biol Chem 2001; 276: 25184–25189.
Reilly JF, Maher PA . Importin beta-mediated nuclear import of fibroblast growth factor receptor: role in cell proliferation. J Cell Biol 2001; 152: 1307–1312.
Peng H, Moffett J, Myers J, Fang X, Stachowiak EK, Maher P et al. Novel nuclear signaling pathway mediates activation of fibroblast growth factor-2 gene by type 1 and type 2 angiotensin II receptors. Mol Biol Cell 2001; 12: 449–462.
Kacinski BM, Scata KA, Carter D, Yee LD, Sapi E, King BL et al. FMS (CSF-1 receptor) and CSF-1 transcripts and protein are expressed by human breast carcinomas in vivo and in vitro. Oncogene 1991; 6: 941–952.
Yee LD, Liu L . The constitutive production of colony stimulating factor 1 by invasive human breast cancer cells. Anticancer Res 2000; 20: 4379–4383.
Yeung YG, Wang Y, Einstein DB, Lee PS, Stanley ER . Colony-stimulating factor-1 stimulates the formation of multimeric cytosolic complexes of signaling proteins and cytoskeletal components in macrophages. J Biol Chem 1998; 273: 17128–17137.
Guilbert LJ, Stanley ER . The interaction of 125I-colony-stimulating factor-1 with bone marrow-derived macrophages. J Biol Chem 1986; 261: 4024–4032.
Hammes LS, Tekmal RR, Naud P, Edelweiss MI, Kirma N, Valente PT et al. Up-regulation of VEGF, c-fms and COX-2 expression correlates with severity of cervical cancer precursor (CIN) lesions and invasive disease. Gynecol Oncol 2008; 110: 445–451.
Zwaenepoel O, Tzenaki N, Vergetaki A, Makrigiannakis A, Vanhaesebroeck B, Papakonstanti EA . Functional CSF-1 receptors are located at the nuclear envelope and activated via the p110delta isoform of PI 3-kinase. FASEB J 2012; 26: 691–706.
Bryant DM, Stow JL . Nuclear translocation of cell-surface receptors: lessons from fibroblast growth factor. Traffic 2005; 10: 947–953.
D'Angelo MA, Hetzer MW . Structure, dynamics and function of nuclear pore complexes. Trends Cell Biol 2008; 18: 456–466.
Brameier M, Krings A, MacCallum RM . NucPred—predicting nuclear localization of proteins. Bioinformatics 2007; 23: 1159–1160.
Offterdinger M, Schofer C, Weipoltshammer K, Grunt TW . c-erbB-3: a nuclear protein in mammary epithelial cells. J Cell Biol 2002; 157: 929–939.
Lin H, Lee E, Hestir K, Leo C, Huang M, Bosch E et al. Discovery of a cytokine and its receptor by functional screening of the extracellular proteome. Science 2008; 320: 807–811.
Shadle PJ, Aldwin L, Nitecki DE, Koths K . Human macrophage colony-stimulating factor heterogeneity results from alternative mRNA splicing, differential glycosylation, and proteolyticprocessing. J Cell Biochem 1989; 40: 91–107.
Pixley FJ, Stanley ER . CSF-1 regulation of the wandering macrophage: complexity in action. Trends Cell Biol 2004; 11: 628–638 Review.
Mancini A, Koch A, Whetton AD, Tamura T . The M-CSF receptor substrate and interacting protein FMIP is governed in its subcellular localization by protein kinase C-mediated phosphorylation, and thereby potentiates M-CSF-mediated differentiation. Oncogene 2004; 39: 6581–6589.
Roussel MF . Signal transduction by the macrophage-colony-stimulating factor receptor (CSF-1R). J Cell Sci Suppl 1994; 18: 105–108.
Dittmann K, Mayer C, Rodemann HP . Nuclear EGFR as novel therapeutic target: insights into nuclear translocation and function. Strahlenther Onkol 2010; 186: 1–6.
Morgan C, Pollard JW, Stanley ER . Isolation and characterization of a cloned growth factor dependent macrophage cell line, BAC1.2F5. J Cell Physiol 1987; 130: 420–427.
Roussel MF, Sherr CJ . Mouse NIH 3T3 cells expressing human colony-stimulating factor 1 (CSF-1) receptors overgrow in serum-free medium containing human CSF-1 as their only growth factor. Proc Natl Acad Sci USA 1989; 86: 7924–7927.
Lehmann BD, Bauer JA, Chen X, Sanders ME, Chakravarthy AB, Shyr Y et al. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J Clin Invest 2011; 121: 2750–2767.
Neve RM, Chin K, Fridlyand J, Yeh J, Baehner FL, Fevr T et al. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell 2006; 10: 515–527.
Stanley ER, Heard PM . Factors regulating macrophage production and growth. Purification and some properties of the colony stimulating factor from medium conditioned by mouse L cells. J Biol Chem 1977; 252: 4305–4312.
Rovida E, Lugli B, Barbetti V, Giuntoli S, Olivotto M, DelloSbarba P . Focal adhesion kinase is redistributed to focal complexes and mediates cell spreading in macrophages in response to M-CSF. Biol Chem 2005; 386: 919–929.
Rovida E, Baccarini M, Olivotto M, DelloSbarba P . Opposite effects of different doses of MCSF on ERK phosphorylation and cell proliferation in macrophages. Oncogene 2002; 21: 3670–3676.
Rovida E, Spinelli E, Sdelci S, Barbetti V, Morandi A, Giuntoli S et al. ERK5/BMK1 is indispensable for optimal colony-stimulating factor 1 (CSF-1)-induced proliferation in macrophages in a Src-dependent fashion. J Immunol 2008; 180: 4166–4172.
Barbetti V, Gozzini A, Rovida E, Morandi A, Spinelli E, Fossati G et al. Selective anti-leukaemic activity of low-dose histone deacetylase inhibitor ITF2357 on AML1/ETO-positive cells. Oncogene 2008; 27: 1767–1778.
Barbetti V, Tusa I, Cipolleschi MG, Rovida E, DelloSbarba P . AML1/ETO sensitizes via TRAIL acute myeloid leukemia cells to the pro-apoptotic effects of hypoxia. Cell Death Dis 2013; 4: e536.
Barbetti V, Gozzini A, Cheloni G, Marzi I, Fabiani E, Santini V et al. Time- and residue-specific differences in histone acetylation induced by VPA and SAHA in AML1/ETO positive leukemia cells. Epigenetics 2013; 8: 210–219.
Acknowledgements
This work was supported by Istituto Toscano Tumori, Associazione Italiana per la Ricerca sul Cancro, Ministero della Salute (Ricerca Finalizzata, Grant number RF-TOS-2008-1163728), Regione Toscana (Programma per la Ricerca in Materia di Salute), Associazione Italiana per la Lotta contro le Leucemie e i Linfomi (sezione di Prato), Fondazione Cassa di Risparmio di Volterra, Fondazione Oretta Bartolomei-Corsi. We thank Professor Clare M Isacke (Breakthrough Breast Cancer Research Centre, London, UK) for revising the manuscript.
Author contributions
ER conceived and designed the experiments. ER, VB, AM, MR, SB, AG, IT, GD, IM and MGC performed the experiments. ER, VB, AM, AG, SB and ADL analyzed the data. ER, AM and PDS wrote the paper.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Oncogene website
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/3.0/
About this article
Cite this article
Barbetti, V., Morandi, A., Tusa, I. et al. Chromatin-associated CSF-1R binds to the promoter of proliferation-related genes in breast cancer cells. Oncogene 33, 4359–4364 (2014). https://doi.org/10.1038/onc.2013.542
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/onc.2013.542
Keywords
This article is cited by
-
Co-axial electrospraying of injectable multi-cancer drugs nanocapsules with polymer shells for targeting aggressive breast cancers
Cancer Nanotechnology (2022)
-
Dynamic gene regulation by nuclear colony-stimulating factor 1 receptor in human monocytes and macrophages
Nature Communications (2019)