Abstract
Post-translational modification (PTM) is an important step of signal transduction that transfers chemical groups such as phosphate, acetyl and glycosyl groups from one protein to another protein. As most of the PTMs are reversible, normal cells use PTMs as a ‘switch’ to determine the resting and proliferating state of cells that enables rapid and tight regulation of cell proliferation. In cancer cells, activation of oncogenes and/or inactivation of tumor suppressor genes provide continuous proliferative signals in part by adjusting the state of diverse PTMs of effector proteins that are involved in regulation of cell survival, cell cycle and proliferation, leading to abnormally fast proliferation of cancer cells. In addition to dysregulated proliferation, ‘altered tumor metabolism’ has recently been recognized as an emerging cancer hallmark. The most common metabolic phenotype of cancer is known as the Warburg effect or aerobic glycolysis that consists of increased glycolysis and enhanced lactate production even in the presence of oxygen. Although Otto Warburg observed aerobic glycolysis nearly 90 years ago, the detailed molecular mechanisms how increased glycolysis is regulated by oncogenic and/or tumor suppressive signaling pathways remain unclear. In this review, we summarize recent advances revealing how these signaling pathways reprogram metabolism through diverse PTMs to provide a metabolic advantage to cancer cells, thereby promoting tumor cell proliferation, tumorigenesis and tumor growth.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout

Similar content being viewed by others
References
Warburg O . On the origin of cancer cells. Science 1956; 123: 309–314.
Hanahan D, Weinberg RA . Hallmarks of cancer: the next generation. Cell 2011; 144: 646–674.
Christofk HR, Vander Heiden MG, Harris MH, Ramanathan A, Gerszten RE, Wei R et al. The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumour growth. Nature 2008; 452: 230–233.
Hitosugi T, Zhou L, Elf S, Fan J, Kang HB, Seo JH et al. Phosphoglycerate mutase 1 coordinates glycolysis and biosynthesis to promote tumor growth. Cancer Cell 2012; 22: 585–600.
Locasale JW, Grassian AR, Melman T, Lyssiotis CA, Mattaini KR, Bass AJ et al. Phosphoglycerate dehydrogenase diverts glycolytic flux and contributes to oncogenesis. Nat Genet 2011; 43: 869–874.
Possemato R, Marks KM, Shaul YD, Pacold ME, Kim D, Birsoy K et al. Functional genomics reveal that the serine synthesis pathway is essential in breast cancer. Nature 2011; 476: 346–350.
Vander Heiden MG, Locasale JW, Swanson KD, Sharfi H, Heffron GJ, Amador-Noguez D et al. Evidence for an alternative glycolytic pathway in rapidly proliferating cells. Science 2010; 329: 1492–1499.
Blume-Jensen P, Hunter T . Oncogenic kinase signalling. Nature 2001; 411: 355–365.
Futreal PA, Coin L, Marshall M, Down T, Hubbard T, Wooster R et al. A census of human cancer genes. Nat Rev Cancer. 2004; 4: 177–183.
Cooper JA, Esch FS, Taylor SS, Hunter T . Phosphorylation sites in enolase and lactate dehydrogenase utilized by tyrosine protein kinases in vivo and in vitro. J Biol Chem 1984; 259: 7835–7841.
Cooper JA, Reiss NA, Schwartz RJ, Hunter T . Three glycolytic enzymes are phosphorylated at tyrosine in cells transformed by Rous sarcoma virus. Nature 1983; 302: 218–223.
Cooper JA, Hunter T . Four different classes of retroviruses induce phosphorylation of tyrosines present in similar cellular proteins. Mol Cell Biol 1981; 1: 394–407.
Medina RA, Owen GI . Glucose transporters: expression, regulation and cancer. Biol Res 2002; 35: 9–26.
Foran PG, Fletcher LM, Oatey PB, Mohammed N, Dolly JO, Tavare JM . Protein kinase B stimulates the translocation of GLUT4 but not GLUT1 or transferrin receptors in 3T3-L1 adipocytes by a pathway involving SNAP-23, synaptobrevin-2, and/or cellubrevin. J Biol Chem 1999; 274: 28087–28095.
Hill MM, Clark SF, Tucker DF, Birnbaum MJ, James DE, Macaulay SL . A role for protein kinase Bbeta/Akt2 in insulin-stimulated GLUT4 translocation in adipocytes. Mol Cell Biol 1999; 19: 7771–7781.
Kohn AD, Summers SA, Birnbaum MJ, Roth RA . Expression of a constitutively active Akt Ser/Thr kinase in 3T3-L1 adipocytes stimulates glucose uptake and glucose transporter 4 translocation. J Biol Chem 1996; 271: 31372–31378.
Kupriyanova TA, Kandror KV . Akt-2 binds to Glut4-containing vesicles and phosphorylates their component proteins in response to insulin. J Biol Chem 1999; 274: 1458–1464.
Tanti JF, Grillo S, Gremeaux T, Coffer PJ, Van Obberghen E, Le Marchand-Brustel Y . Potential role of protein kinase B in glucose transporter 4 translocation in adipocytes. Endocrinology 1997; 138: 2005–2010.
Robey RB, Hay N . Is Akt the "Warburg kinase"?-Akt-energy metabolism interactions and oncogenesis. Sem Cancer Biol 2009; 19: 25–31.
Plas DR, Talapatra S, Edinger AL, Rathmell JC, Thompson CB . Akt and Bcl-xL promote growth factor-independent survival through distinct effects on mitochondrial physiology. J Biol Chem 2001; 276: 12041–12048.
Rathmell JC, Fox CJ, Plas DR, Hammerman PS, Cinalli RM, Thompson CB . Akt-directed glucose metabolism can prevent Bax conformation change and promote growth factor-independent survival. Mol Cell Biol 2003; 23: 7315–7328.
Elstrom RL, Bauer DE, Buzzai M, Karnauskas R, Harris MH, Plas DR et al. Akt stimulates aerobic glycolysis in cancer cells. Cancer Res 2004; 64: 3892–3899.
Gottlob K, Majewski N, Kennedy S, Kandel E, Robey RB, Hay N . Inhibition of early apoptotic events by Akt/PKB is dependent on the first committed step of glycolysis and mitochondrial hexokinase. Genes Dev 2001; 15: 1406–1418.
Miyamoto S, Murphy AN, Brown JH . Akt mediates mitochondrial protection in cardiomyocytes through phosphorylation of mitochondrial hexokinase-II. Cell Death Diff 2008; 15: 521–529.
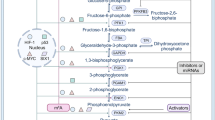
Deprez J, Vertommen D, Alessi DR, Hue L, Rider MH . Phosphorylation and activation of heart 6-phosphofructo-2-kinase by protein kinase B and other protein kinases of the insulin signaling cascades. J Biol Chem 1997; 272: 17269–17275.
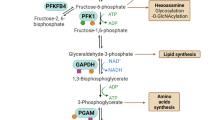
Yi W, Clark PM, Mason DE, Keenan MC, Hill C, Goddard WA 3rd et al. Phosphofructokinase 1 glycosylation regulates cell growth and metabolism. Science 2012; 337: 975–980.
Britton HG, Clarke JB . Mechanism of the 2,3-diphosphoglycerate-dependent phosphoglycerate mutase from rabbit muscle. Biochemical J 1972; 130: 397–410.
Fothergill-Gilmore LA, Watson HC . The phosphoglycerate mutases. Adv Enzymol Relat Areas Mol Biol 1989; 62: 227–313.
Grisolia S, Cleland WW . Influence of salt, substrate, and cofactor concentrations on the kinetic and mechanistic behavior of phosphoglycerate mutase. Biochemistry 1968; 7: 1115–1121.
Rose ZB, Dube S . Rates of phosphorylation and dephosphorylation of phosphoglycerate mutase and bisphosphoglycerate synthase. J Biol Chem 1976; 251: 4817–4822.
Rose ZB, Hamasaki N, Dube S . The sequence of a peptide containing the active site phosphohistidine residue of phosphoglycerate mutase from chicken breast muscle. J Biol Chem 1975; 250: 7939–7942.
Corcoran CA, Huang Y, Sheikh MS . The regulation of energy generating metabolic pathways by p53. Cancer Biol Ther 2006; 5: 1610–1613.
Tennant DA, Duran RV, Boulahbel H, Gottlieb E . Metabolic transformation in cancer. Carcinogenesis 2009; 30: 1269–1280.
Tennant DA, Duran RV, Gottlieb E . Targeting metabolic transformation for cancer therapy. Nat Rev Cancer 2010; 10: 267–277.
Hitosugi T, Zhou L, Fan J, Elf S, Zhang L, Xie J et al. Tyr26 phosphorylation of PGAM1 provides a metabolic advantage to tumours by stabilizing the active conformation. Nat Commun 2013; 4: 1790.
Hallows WC, Yu W, Denu JM . Regulation of glycolytic enzyme phosphoglycerate mutase-1 by Sirt1 protein-mediated deacetylation. J Biol Chem 2012; 287: 3850–3858.
Evans MJ, Morris GM, Wu J, Olson AJ, Sorensen EJ, Cravatt BF . Mechanistic and structural requirements for active site labeling of phosphoglycerate mutase by spiroepoxides. Mol Biosyst 2007; 3: 495–506.
Evans MJ, Saghatelian A, Sorensen EJ, Cravatt BF . Target discovery in small-molecule cell-based screens by in situ proteome reactivity profiling. Nat Biotechnol 2005; 23: 1303–1307.
Christofk HR, Vander Heiden MG, Wu N, Asara JM, Cantley LC . Pyruvate kinase M2 is a phosphotyrosine-binding protein. Nature 2008; 452: 181–186.
Hitosugi T, Kang S, Vander Heiden MG, Chung TW, Elf S, Lythgoe K et al. Tyrosine phosphorylation inhibits PKM2 to promote the Warburg effect and tumor growth. Sci Signaling 2009; 2: ra73.
Dang CV . PKM2 tyrosine phosphorylation and glutamine metabolism signal a different view of the Warburg effect. Sci Signaling 2009; 2: pe75.
Gao X, Wang H, Yang JJ, Liu X, Liu ZR . Pyruvate kinase M2 regulates gene transcription by acting as a protein kinase. Mol Cell 2012; 45: 598–609.
Hoshino A, Hirst JA, Fujii H . Regulation of cell proliferation by interleukin-3-induced nuclear translocation of pyruvate kinase. J Biol Chem 2007; 282: 17706–17711.
Luo W, Hu H, Chang R, Zhong J, Knabel M, O'Meally R et al. Pyruvate kinase M2 is a PHD3-stimulated coactivator for hypoxia-inducible factor 1. Cell 2011; 145: 732–744.
Stetak A, Veress R, Ovadi J, Csermely P, Keri G, Ullrich A . Nuclear translocation of the tumor marker pyruvate kinase M2 induces programmed cell death. Cancer Res 2007; 67: 1602–1608.
Yang W, Xia Y, Hawke D, Li X, Liang J, Xing D et al. PKM2 phosphorylates histone H3 and promotes gene transcription and tumorigenesis. Cell 2012; 150: 685–696.
Yang W, Zheng Y, Xia Y, Ji H, Chen X, Guo F et al. ERK1/2-dependent phosphorylation and nuclear translocation of PKM2 promotes the Warburg effect. Nat Cell Biol 2012; 14: 1295–1304.
Yang W, Xia Y, Ji HT, Zheng YH, Liang J, Huang WH et al. Nuclear PKM2 regulates beta-catenin transactivation upon EGFR activation. Nature 2011; 480: 118–U289.
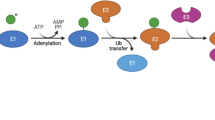
Lv L, Li D, Zhao D, Lin R, Chu Y, Zhang H et al. Acetylation targets the M2 isoform of pyruvate kinase for degradation through chaperone-mediated autophagy and promotes tumor growth. Mol Cell 2011; 42: 719–730.
Anastasiou D, Poulogiannis G, Asara JM, Boxer MB, Jiang JK, Shen M et al. Inhibition of pyruvate kinase M2 by reactive oxygen species contributes to cellular antioxidant responses. Science 2011; 334: 1278–1283.
Anastasiou D, Yu Y, Israelsen WJ, Jiang JK, Boxer MB, Hong BS et al. Pyruvate kinase M2 activators promote tetramer formation and suppress tumorigenesis. Nat Chem Biol 2012; 8: 839–847.
Parnell KM, Foulks JM, Nix RN, Clifford A, Bullough J, Luo B et al. Pharmacologic activation of PKM2 slows lung tumor xenograft growth. MolCancer Ther 2013; 12: 1453–1460.
Fan J, Hitosugi T, Chung TW, Xie J, Ge Q, Gu TL et al. Tyrosine phosphorylation of lactate dehydrogenase A is important for NADH/NAD(+) redox homeostasis in cancer cells. Mol Cell Biol 2011; 31: 4938–4950.
Zhao D, Zou SW, Liu Y, Zhou X, Mo Y, Wang P et al. Lysine-5 acetylation negatively regulates lactate dehydrogenase a and is decreased in pancreatic cancer. Cancer Cell 2013; 23: 464–476.
Kim HS, Vassilopoulos A, Wang RH, Lahusen T, Xiao Z, Xu X et al. SIRT2 maintains genome integrity and suppresses tumorigenesis through regulating APC/C activity. Cancer Cell 2011; 20: 487–499.
Patel MS, Korotchkina LG . Regulation of mammalian pyruvate dehydrogenase complex by phosphorylation: complexity of multiple phosphorylation sites and kinases. Exp Mol Med 2001; 33: 191–197.
Roche TE, Hiromasa Y . Pyruvate dehydrogenase kinase regulatory mechanisms and inhibition in treating diabetes, heart ischemia, and cancer. Cell Mol Life Sci 2007; 64: 830–849.
Hitosugi T, Fan J, Chung TW, Lythgoe K, Wang X, Xie J et al. Tyrosine phosphorylation of mitochondrial pyruvate dehydrogenase kinase 1 is important for cancer metabolism. Mol Cell 2011; 44: 864–877.
Bonnet S, Archer SL, Allalunis-Turner J, Haromy A, Beaulieu C, Thompson R et al. A mitochondria-K+ channel axis is suppressed in cancer and its normalization promotes apoptosis and inhibits cancer growth. Cancer Cell 2007; 11: 37–51.
Michelakis ED, Sutendra G, Dromparis P, Webster L, Haromy A, Niven E et al. Metabolic modulation of glioblastoma with dichloroacetate. Sci Transl Med 2010; 2: 31ra4.
Brahimi-Horn MC, Chiche J, Pouyssegur J . Hypoxia signalling controls metabolic demand. Curr Opi Cell Biol 2007; 19: 223–229.
Cheung EC, Vousden KH . The role of p53 in glucose metabolism. Curr Opi Cell Biol 2010; 22: 186–191.
Gordan JD, Thompson CB, Simon MC . HIF and c-Myc: sibling rivals for control of cancer cell metabolism and proliferation. Cancer Cell 2007; 12: 108–113.
Kondoh H, Lleonart ME, Gil J, Wang J, Degan P, Peters G et al. Glycolytic enzymes can modulate cellular life span. Cancer Res 2005; 65: 177–185.
Kroemer G, Pouyssegur J . Tumor cell metabolism: cancer's Achilles' heel. Cancer Cell 2008; 13: 472–482.
Semenza GL, Roth PH, Fang HM, Wang GL . Transcriptional regulation of genes encoding glycolytic enzymes by hypoxia-inducible factor 1. J Biol Chem 1994; 269: 23757–23763.
Shim H, Dolde C, Lewis BC, Wu CS, Dang G, Jungmann RA et al. c-Myc transactivation of LDH-A: implications for tumor metabolism and growth. Proc Natl Acad Sci USA 1997; 94: 6658–6663.
Hara MR, Agrawal N, Kim SF, Cascio MB, Fujimuro M, Ozeki Y et al. S-nitrosylated GAPDH initiates apoptotic cell death by nuclear translocation following Siah1 binding. Nat Cell Biol 2005; 7: 665–674.
Doskeland AP, Flatmark T . Recombinant human phenylalanine hydroxylase is a substrate for the ubiquitin-conjugating enzyme system. Biochem J 1996; 319 (Pt 3): 941–945.
Chouchani ET, James AM, Fearnley IM, Lilley KS, Murphy MP . Proteomic approaches to the characterization of protein thiol modification. Curr Opin Chem Biol 2011; 15: 120–128.
Morelle W, Canis K, Chirat F, Faid V, Michalski JC . The use of mass spectrometry for the proteomic analysis of glycosylation. Proteomics 2006; 6: 3993–4015.
Acknowledgements
We apologize to authors whose contributions were not directly cited owing to space limitations. This study is supported in part by NIH grants CA140515 (JC) and DoD grant W81XWH-12–1–0217 (JC). JC is a Georgia Cancer Coalition Distinguished Cancer Scholar and a Scholar of the Leukemia and Lymphoma Society.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Rights and permissions
About this article
Cite this article
Hitosugi, T., Chen, J. Post-translational modifications and the Warburg effect. Oncogene 33, 4279–4285 (2014). https://doi.org/10.1038/onc.2013.406
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/onc.2013.406
Keywords
This article is cited by
-
PRMT2 silencing regulates macrophage polarization through activation of STAT1 or inhibition of STAT6
BMC Immunology (2024)
-
PIGT promotes cell growth, glycolysis, and metastasis in bladder cancer by modulating GLUT1 glycosylation and membrane trafficking
Journal of Translational Medicine (2024)
-
Transcriptional regulation and post-translational modifications in the glycolytic pathway for targeted cancer therapy
Acta Pharmacologica Sinica (2024)
-
Protein post-translational modifications in the regulation of cancer hallmarks
Cancer Gene Therapy (2023)
-
Regulation of tumor metabolism by post translational modifications on metabolic enzymes
Cancer Gene Therapy (2023)