Abstract
Estrogen receptor α (ERα) has critical roles in the development and progression of breast cancer, and the coiled-coil co-activator (CoCoA) is an important ERα co-activator for estrogen-induced gene expression. The small ubiquitin-like modifier (SUMO) pathway is hyperactivated in breast cancer, but the mechanism by which SUMOylation regulates ERα-mediated transcription remains poorly understood. Here, we identified ZFP282 as a CoCoA-binding protein. ZFP282 associates directly with ERα and cooperates synergistically with CoCoA to enhance ERα function. ZFP282 is required for estrogen-induced expression of ERα target genes and estrogen-dependent breast cancer cell growth and tumorigenesis. In addition, we found that ZFP282 is SUMOylated and that SUMOylation positively regulates the co-activator activity of ZFP282 by increasing its binding affinity to ERα and CoCoA, and consequently increasing recruitment of ZFP282–CoCoA complex to the promoter of ERα target genes. These findings reveal essential roles for ZFP282 and its SUMOylation in estrogen signaling and breast tumorigenesis.
Similar content being viewed by others
Introduction
Post-translational modifications have a key role in the functional regulation of transcription factors and their co-regulators. Among various post-translational modifications, SUMOylation has recently been identified as an important mechanism to regulate numerous cellular processes.1 SUMOylation is a covalent modification that adds small ubiquitin-like modifier (SUMO) to target substrates. SUMOylation is a multistep process involving the heterodimeric SUMO-activating enzyme SAE1/SAE2 (E1), the SUMO-conjugating enzyme Ubc9 (E2), and in some cases, the SUMO E3 ligases. SUMOylation mostly occurs on the lysine residue within a ψKXE consensus motif (where ψ is a large hydrophobic amino acid and X is any amino acid), and the modification can be dynamically reversed by the sentrin-specific proteases (SENPs).1 SUMOylation has been found to regulate protein function in a variety of ways, including effects on subcellular localization, protein–protein interactions, protein stability and transcriptional activity. Because many transcription factors and their co-regulators are subjected to SUMO modification, SUMOylation has emerged as an important mechanism in controlling gene expression.2 Although SUMOylation has been associated with both activation and repression of transcriptional activities, a large majority of the reports published so far have shown repressive effects of SUMOylation on transcription.2
Estrogen receptor α (ERα) is a member of the nuclear receptor (NR) superfamily, and it functions as a ligand-dependent transcription factor that mediates the diverse biological effects of estrogens, including development, reproduction and the etiology of breast cancer.3 The p160 transcriptional co-activators, SRC-1, GRIP1/SRC-2 and AIB1/SRC-3, bind directly to NRs and serve as protein scaffolds for the assembly of multicomponent co-activator complexes on target promoters.4 p160 co-activators recruit secondary co-activators, including histone acetyltransferase p300/CBP, histone methyltransferase CARM1 and coiled-coil co-activator (CoCoA), and act synergistically with secondary co-activators to enhance NR function.5, 6 CoCoA interacts indirectly with NRs through the p160 co-activators. CoCoA consists of three functional domains: the central coiled-coil domain, which interacts with the bHLH-PAS domains of p160 co-activators; the C-terminal activation domain (AD); and the N-terminal AD (Figure 1a). The C-terminal AD of CoCoA is essential for its co-activator function with NRs and contributes to transcriptional activation through multiple protein–protein interactions with p300, CCAR1 and DBC1.6, 7, 8 To further elucidate the molecular role of the CoCoA C-terminal AD, we performed a yeast two-hybrid screen using the C-terminal AD of CoCoA as bait and found a Krüppel-type zinc finger protein, ZFP282 (also known as HUB1), as a CoCoA-binding protein. ZFP282 was originally identified as a human T-cell leukemia virus type I (HTLV-I) U5 repressive element (U5RE)-binding protein.9 Here we show that ZFP282 associates and cooperates synergistically with CoCoA to enhance ERα function. ZFP282 is required for the expression of ERα target genes and estrogen-dependent breast cancer cell growth and tumorigenesis. In addition, we found that ZFP282 is SUMOylated and that SUMOylation positively regulates the co-activator activity of ZFP282 by increasing its binding affinity to ERα and CoCoA.
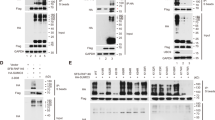
ZFP282 interacts with CoCoA and ERα. (a, b) Schematic representation of CoCoA and ZFP282 structures. AD, activation domain; Bait, amino acids 661–691 of CoCoA; KRAB, Krüppel-associated box; IR, internal repeat; ψKXE, SUMOylation consensus motif. (c) COS-7 cells were transfected with pSG5.HA-CoCoA and pFLAG.CMV4-ZFP282 or empty vector pFLAG.CMV4. Cell lysates were immunoprecipitated with anti-FLAG antibody or control normal IgG. Immunoprecipitated ZFP282 and coimmunoprecipitated CoCoA were detected by anti-FLAG antibody and anti-HA antibody, respectively. (d) In vitro translated HA-tagged CoCoA and its fragments were incubated with recombinant GST-ZFP282 bound to beads. Bound proteins were analyzed by immunoblot with anti-HA antibody. (e) COS-7 cells were transfected with pSG5.HA-ERα and pFLAG.CMV4-ZFP282 or empty vector and treated with ethanol or 100 nM E2 for 1 h. CoIP experiments were performed, as described in (c). (f) In vitro translated HA-tagged ZFP282 was incubated with recombinant GST-ERα LBD in the presence or absence of E2. Bound proteins were analyzed by immunoblot with anti-HA antibody.
Results
Isolation of ZFP282 as a CoCoA-binding protein
To further understand the function of CoCoA AD and to identify its binding partners, we performed a yeast two-hybrid screen of a mouse 17 day embryo cDNA library using amino acids 661–691 as bait (Figure 1a) and isolated a cDNA clone encoding zinc finger protein 282 (ZFP282) from several rounds of screening and selection. Full-length mouse ZFP282 cDNA encodes a protein of 667 amino acids (Figure 1b). ZFP282 contains five tandem-repeated C2H2-type zinc fingers at the C-terminus and a Krüppel-associated box domain at the N-terminus. The central domain of ZFP282 contains three internal repeats (IR) of 40 amino acids and a glycine-rich domain.
The interaction between CoCoA and ZFP282 was confirmed by coimmunoprecipitation (CoIP) and glutathione-S-transferase (GST) pull-down assays. In CoIP experiments, FLAG-tagged ZFP282 was coimmunoprecipitated specifically with HA-tagged CoCoA from extracts of transiently transfected COS-7 cells (Figure 1c). GST-ZFP282 interacted with full-length CoCoA and its C-terminal domain (Figure 1d and Supplementary Figure S1). The N-terminal, but not the central coiled-coil, region of CoCoA was also capable of binding to ZFP282. Thus, CoCoA has two independent ZFP282-binding sites. Because CoCoA-interacting proteins, including p160 co-activators, CCAR1 and DBC1, can interact with NRs, we also tested whether ZFP282 binds to ERα. A specific interaction between ERα and ZFP282 was detected by CoIP and GST pull-down assays, and the interaction was increased modestly by estradiol (E2) (Figures 1e and f).
ZFP282 functions as an ERα co-activator and cooperates synergistically with CoCoA
To test whether ZFP282 can function as a co-activator for ERα, we transfected CV-1 cells with an ERα reporter plasmid along with expression vectors for ERα and ZFP282. ZFP282 enhanced ERα function in a dose-dependent and hormone-dependent manner (Figure 2a). ZFP282 also enhanced transcriptional activation by AR, GR and TR in a hormone-dependent manner (Supplementary Figure S2). When ZFP282 was coexpressed with CoCoA, a synergistic induction of ERα-mediated transactivation was observed (Figure 2b). These results suggest that ZFP282 functions as a co-activator for multiple NRs and cooperates with CoCoA to enhance NR-dependent transcriptional activation. As CoCoA alone did not enhance ERα-mediated transcription and as CoCoA does not bind to ERα,5 the co-activator synergy between CoCoA and ZFP282 suggests that ZFP282 recruits CoCoA to the reporter gene through its association with ERα. ZFP282 also stimulated the activity of CoCoA C-terminal AD fused to Gal4-DNA-binding domain), suggesting the functional interaction between ZFP282 and CoCoA (Figure 2c).
ZFP282 functions as an ERα co-activator and cooperates synergistically with CoCoA. (a) CV-1 cells were transfected with pHE0 encoding ERα (2 ng) and 2ERE-TATA-LUC reporter in combination with various amounts (100, 200 and 400 ng) of pSG5.HA-ZFP282 and grown in medium containing or lacking 100 nM E2 before conducting luciferase assays on cell extracts. Data are means±s.d. (n=3). (b) Transient transfections were performed, as described in (a). Expression vectors: pSG5.HA-ZFP282 (200 ng), pSG5.HA-CoCoA (200 ng). Data are means±s.d. (n=3). (c) CV-1 cells were transfected with pG5-LUC reporter (200 ng), pSG5.HA-ZFP282 (200 and 400 ng), and plasmids encoding Gal4-DNA-binding domain or Gal4-CoCoA 470–691 (200 ng), as indicated and harvested for luciferase assays. Data are means±s.d. (n=3). (d) ChIP assay. Crosslinked, sheared chromatin from MCF-7 cells treated with or without 10 nM E2 (45 min) was immunoprecipitated with the indicated antibodies. Quantitative PCR (qPCR) analyses were performed using primers specific for the pS2 promoter. The results are shown as percentage of input and are means±s.d. (n=3). (e) ChIP and ReIP assay, using the indicated antibodies, were performed, as described in (d).
To examine whether ZFP282 is directly involved in ERα-mediated transcription, we performed chromatin immunoprecipitation (ChIP) assays in MCF-7 cells (Figure 2d). ERα, AIB1 and CoCoA were recruited to the ERα-binding site associated with the pS2 gene promoter in a hormone-dependent manner, as reported previously.5, 7, 8 E2 treatment also led to the recruitment of ZFP282 to the pS2 promoter, whereas the normal IgG control was not affected by E2. A similar recruitment pattern of ZFP282 was observed on ERα-binding sites associated with the promoters of other ERα target genes, CTSD, GREB1 and PgR (Supplementary Figure S3), indicating ZFP282 is directly involved in the transcriptional regulation of endogenous ERα target genes. ChIP experiments with sequential immunoprecipitation using anti-ZFP282 and anti-CoCoA antibodies demonstrated that ZFP282 and CoCoA exist in the same complex on the pS2 promoter (Figure 2e). Thus, E2 induces ZFP282–CoCoA complex formation on the pS2 promoter.
A previous study reported that ZFP282 binds U5RE of HTLV-1 long terminal repeat (LTR) and regulates HTLV-1 LTR-mediated gene expression.9 Thus, we next examined the effect of ZFP282 on U5RE-mediated transcription. ZFP282 strongly enhanced the transcriptional activity of a reporter construct containing five copies of the U5RE in a dose-dependent manner (Supplementary Figure S4a), and coexpression of CoCoA further enhanced the reporter activity (Supplementary Figure S4b). Similar results were observed in reporter assays using a reporter containing natural HTLV-1 LTR (Supplementary Figure S4), suggesting that ZFP282 functions as a transcriptional activator of HTLV-1 LTR-driven reporter gene expression.
ZFP282 is required for E2-induced expression of ERα target genes
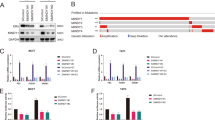
To further assess the functional involvement of ZFP282 in ERα-mediated transcription, the expression of ZFP282 was reduced by RNA interference. To avoid possible off-target effects caused by the ZFP282 RNA interference, we used two different small interefering RNA sequences that target different regions of ZFP282 mRNA. When ZFP282 mRNA and protein levels were specifically reduced in MCF-7 cells by small interefering RNA transfection (Figure 3a), E2-induced expression of the endogenous pS2, GREB1, PgR and CTSD genes was significantly inhibited compared with the results using nonspecific (NS) small interefering RNA (Figure 3b). These results are consistent with the ChIP data demonstrating E2-induced ZFP282 recruitment to each of these gene promoters and strongly suggest that ZFP282 is directly involved in E2 induction of ERα target gene expression. Similarly, depletion of ZFP282 also repressed E2-induced expression of a transiently transfected reporter gene and transcriptional activity of Gal4-CoCoA AD in MCF-7 cells (Figures 3c and d), suggesting that ZFP282 is required for both E2-induced expression of ERα target genes and full transcriptional activity of CoCoA AD.
ZFP282 is required for E2-induced expression of ERα target genes. (a, b) MCF-7 cells were transfected with 40 pmol of ZFP282 siRNA#5, #9 or NS siRNA duplex. 72 h after transfection, cells were treated or untreated with 10 nM E2 and harvested after an additional 24 h. Total RNA was examined by real-time quantitative RT–PCR (qRT–PCR) analysis with primers specific for the indicated mRNAs. Results shown were normalized to β-actin mRNA levels and are means±s.d. (n=3). Protein levels were monitored by immunoblot using the indicated antibodies. (c) MCF-7 cells were transfected with 2ERE-TK-LUC and 40 pmol of either the siZFP282#5 or siNS, treated or untreated with E2 as in (a), and harvested for luciferase assays. Data are means±s.d. (n=3). (d) MCF-7 cells were transfected with pG5-LUC, plasmids encoding Gal4 or Gal4-CoCoA 470–691, and 40 pmol of either the siZFP282#5 or siNS, as indicated and harvested for luciferase assays. Data are means±s.d. (n=3).
ZFP282 is required for tumorigenic potential of breast cancer cells
To assess the potential relevance of our results to the biology of breast cancer cells, we examined the effect of reduced ZFP282 levels on E2-stimulated cell proliferation and cell cycle progression in MCF-7 cells transfected with a siNS or ZFP282 small interefering RNA (Figure 4a). E2 treatment stimulated proliferation and cell cycle progression of MCF-7/siNS cells, but E2 stimulation of MCF-7 cell growth and cell cycle progression was attenuated by ZFP282 depletion (Figures 4b and c). The percentage of S-phase cells was reduced from 28 to 17%. Because ERα mediates estrogen-induced G1/S transition and cell proliferation, these results suggest that ZFP282 has a critical role as an ERα co-activator in E2-stimulated proliferation of breast cancer cells and in promoting cell cycle progression. We next examined the effect of ZFP282 depletion on migration, colony formation and invasion in MCF-7 cells infected with lentiviruses expressing a shNS or ZFP282 shRNA (shZFP282) (Figure 4a). Depletion of ZFP282 decreased migration and clonogenic survival of MCF-7 cells and inhibited MCF-7 cell invasion (Figures 4d–f), suggesting that ZFP282 is also required for cellular properties associated with the transformed phenotype of breast cancer cells. To further investigate the role of ZFP282 in tumorigenic potential of breast cancer cells, we assessed the effect of ZFP282 depletion on the growth of MCF-7 xenograft tumors in nude mice injected with ZFP282 knockdown (shZFP282) cells or control (shNS) cells (Figure 4g). As expected,10 growth of MCF-7 xenograft tumors was strictly E2-dependent. MCF-7/shNS tumors grew rapidly in E2-treated mice; in contrast, E2-stimulated tumor growth was almost abolished in mice injected with MCF-7/shZFP282 cells. Taken together, these results strongly suggest that ZFP282 has a pivotal role in tumorigenic growth of breast cancer cells.
ZFP282 is required for breast cancer cell proliferation and tumorigenesis. (a) Depletion of ZFP282 expression. MCF-7 cells were transfected with siRNAs, as indicated or infected with lentiviruses encoding a NS or ZFP282 shRNA. Protein levels were monitored by immunoblot using the indicated antibodies. (b) Proliferation assay. MCF-7 cells transfected with indicated siRNAs were cultured in the absence or presence of 1 nM E2 for 8 days. Cell viability was determined by MTT assay. Data are means±s.d. (n=6). P value was determined by Student’s t-test. (c) Cell cycle analysis. MCF-7 cells transfected with the indicated siRNAs were cultured in the absence or presence of 10 nM E2 for 24 h and subjected to fluorescence-activated cell sorter (FACS) analysis. Data are means±s.d. (n=4). P value was determined by Student’s t-test (*P<0.05). (d–f) Migration, colony formation and invasion assays. MCF-7 cells were infected with lentiviruses encoding a NS or ZFP282 shRNA. Representative images of cell migration, anchorage-dependent colony formation and invasion assays are shown on the left panel of each figure, and the quantitative analysis (n=4, ±s.d.) is shown on the right panel of each figure. Scratch assay was performed on confluent cell monolayers, and the white line indicates the migration front of cells (d). For colony-formation assay, viable colonies were stained with crystal violet; after washing, the dye was extracted with 10% SDS solution and quantified by absorbance at 570 nm (e). Invasion assay was performed using a modified Boyden chamber, as described under Supplementary Information (f). (g) Effect of ZFP282 depletion on the growth of MCF-7 xenograft tumors. MCF-7 cells infected with the indicated lentiviral shRNA vectors were injected subcutaneously in athymic female mice. Tumor growth was monitored for 10 weeks under the treatment with vehicle or E2 in drinking water. Representative images of tumor-bearing mice and their tumors are shown on the left panel and the average (n=4, ±s.d.) was shown on the right panel. P value was determined by Student’s t-test (*P<0.05).
ZFP282 is modified by SUMO
Considering that sumoylation is often associated with a change in the activity of transcriptional co-regulators2, 11 and that ZFP282 harbors three SUMOylation consensus motifs (ψKXE) in the central IR region (Figure 1b), we tested whether ZFP282 could be SUMOylated. COS-7 cells were transfected with plasmids expressing HA-tagged ZFP282 and SUMO1 fused to EGFP, and the whole-cell extracts were immunoblotted with anti-HA antibody (Figure 5a). Coexpression of HA-tagged ZFP282 with EGFP-SUMO1 produced multiple additional bands migrating above the unmodified ZFP282, suggesting that ZFP282 is SUMOylated at multiple sites. Immunoprecipitation of cell lysates with anti-HA antibody followed by immunoblotting with anti-SUMO1 verified that the slower migrating bands indeed corresponded to ZFP282 covalently modified by SUMO1 (Figure 5b). Using a bacterial sumoylation system, we confirmed that ZFP282 is a target for SUMO modification (Figure 5c). We next determined the effect of SUMO pathway components such as Ubc9 and SENP1 on ZFP282 SUMOylation. ZFP282 bound to Ubc9 (Supplementary Figure S5a), and expression of Ubc9 robustly increased the level of SUMOylated ZFP282 (Supplementary Figure S5b). Expression of SENP1, a SUMO-specific protease, decreased the amount of SUMOylated ZFP282 forms, whereas a catalytically inactive SENP1 mutant did not affect their amount (Supplementary Figure S5c).
ZFP282 is covalently modified by SUMO. (a, b) In vivo SUMOylation assay. COS-7 cells were transfected with pSG5.HA-ZFP282 and a plasmid encoding EGFP-SUMO1 as indicated. Cell extracts were prepared in the presence of N-ethylmaleimide and analyzed by immunoblot with anti-HA antibody (a) or by immunoprecipitation and immunoblot with the indicated antibodies (b). (c) SUMOylation of ZFP282 in E. coli. GST-ZFP282 was expressed in E. coli in the presence or absence of pT-E1E2S1 plasmid encoding Aos1/Uba2, Ubc9, and SUMO1. The purified ZFP282 proteins were analyzed by SDS–PAGE followed by Coomassie blue staining. (d) COS-7 cells were transfected with pSG5.HA-ZFP282 deletion mutants and a plasmid encoding EGFP-SUMO1 as indicated, and immunoblot was performed as in (a). (e) COS-7 cells were transfected with a plasmid encoding EGFP-SUMO1 and pSG5.HA-ZFP282 WT or various KR mutants as indicated, and immunoblot was performed as in (a).
Because the central IR region harbors three SUMOylation consensus motifs at amino acids 293, 331 and 369 and because the major SUMOylation sites are mapped to the IR region (Figure 5d), we generated ZFP282 mutants in which lysine residues in the consensus motifs were changed to arginines individually or in combination (2KR or 3KR). Three single-point mutations led to significant decrease of the highest migrating band, and the second band above the unmodified ZFP282 was strongly reduced by double mutation (Figure 5e). The triple K293/331/369R mutation (3KR) completely abolished all SUMO-modified forms of ZFP282. Taken together, these findings indicate that K293, K331 and K369 are major SUMO acceptor sites in ZFP282. Notably, these consensus sites are highly conserved in mammals, suggesting functional importance (Supplementary Figure S6).
SUMOylation of ZFP282 positively regulates its co-activator activity and enhances estrogen-stimulated breast cancer cell growth
SUMOylation can affect the activity and subcellular localization of target proteins.1 In immunofluorescence confocal microscopy experiments, ZFP282 wild type (WT) was located mainly in the nucleus, and no difference in the cellular distribution of ZFP282 WT and SUMOylation-defective 3KR mutant was observed (Supplementary Figure S7). These results suggest that SUMOylation of ZFP282 is not likely involved in altering its intracellular localization. To gain an insight into the role of the SUMOylation of ZFP282, we next examined the effects of SUMOylation on the co-activator activity of ZFP282. In ERα reporter gene assays, the 3KR mutant displayed much reduced co-activator activity and synergy with CoCoA compared with those of ZFP282 WT (Figures 6a and b). We next examined whether SUMOylated ZFP282 is present on the pS2 promoter. Sequential ChIP experiments with serial immunoprecipitation using anti-ZFP282 and anti-SUMO1 antibodies demonstrated that ZFP282–SUMO1 complex was recruited to the pS2 promoter under active conditions (Figure 6c). Although the possibility that other SUMOylated proteins bind to ZFP282 on the pS2 promoter cannot be excluded, it is possible that SUMOylated ZFP282 exists on the pS2 promoter. Together, these results suggest that the SUMOylation target lysines are required for the co-activator activity of ZFP282 and synergy with CoCoA. Similar results were observed in reporter gene assays and ChIP assays with the 5XU5RE-TA-LUC construct (Supplementary Figures S8 and S9).
SUMOylation of ZFP282 positively regulates its co-activator activity and enhances hormone-stimulated breast cancer cell growth. (a) Transient transfection in CV-1 cells using 2ERE-TATA-LUC and expression vectors for ERα, HA-ZFP282 or HA-ZFP282 3KR (200 and 400 ng) were performed, as described in Figure 2a. Data are means±s.d. (n=3). Expression levels of transfected ZFP282 were analyzed by immunoblot with anti-HA antibody. (b) Transient transfection was performed as in (a) with the additional plasmid pSG5.HA-CoCoA (200 ng). Data are means±s.d. (n=3). (c) ChIP and ReIP assay, using the indicated antibodies, was performed, as described in Figure 2e. Data are means±s.d. (n=3). (d) Transient transfection was performed as in (a) using pSG5.HA constructs encoding ZFP282 or SUMO1-fused ZFP282 as indicated. Data are means±s.d. (n=3). Expression levels of transfected ZFP282 were analyzed by immunoblot with anti-HA antibody. (e) Cell proliferation assays were performed, as described in Figure 4b using MCF-7 cells transfected with pSG5.HA-ZFP282 plasmids (WT, 3KR, or SUMO1-fused ZFP282 3KR). Data are means±s.d. (n=6). *P<0.05 and **P<0.01.
To further confirm the functional roles of ZFP282 SUMOylation, we next tested the effect of ZFP282 SUMOylation-mimetic mutant on ERα-mediated transcription. To this end, we fused SUMO1 to the N-terminus of ZFP282 to generate a constitutively SUMOylated form of ZFP282 and to eliminate the complication of the indirect effects associated with overexpression of SUMO1 or Ubc9. This approach has been successfully used to characterize the function of SUMOylation of CoCoA.12 In ERα reporter gene assays, expression of SUMO1-fused ZP282 more strongly enhanced the ERα-mediated transcription compared with that of ZFP282 WT (Figure 6d). Similarly, SUMO1 fusion partially restored co-activator activity of ZFP282 3KR mutant. The same pattern of SUMOylation effects of ZFP282 was observed in reporter gene assays with the 5XU5RE-LUC construct (Supplementary Figure S8c). In MTT assays, expression of ZFP282 enhanced E2-stimulated MCF-7 cell proliferation much more efficiently than ZFP282 3KR mutant (Figure 6e). Importantly, SUMO1-fused ZFP282 3KR mutant partially restored the ability of ZFP282 to enhance E2-stimulated cell proliferation. These results suggest that SUMOylation may be an active mechanism regulating the co-activator function of ZFP282 and that SUMOylation of ZFP282 is required for the enhancement of E2-stimulated breast cancer cell growth.
SUMOylation of ZFP282 increases its interaction with CoCoA and ERα
The above observations prompted us to examine whether SUMOylation of ZFP282 could affect the interaction with its binding partners, CoCoA and ERα. In CoIP experiments, ZFP282 WT was efficiently coimmunoprecipitated with CoCoA, whereas a small amount of ZFP282 3KR was precipitated by CoCoA, suggesting that SUMOylation of ZFP282 may enhance CoCoA interaction (Figure 7a). We next examined the interaction between endogenous ZFP282 and CoCoA in the presence or absence of N-ethylmaleimide, which inhibits deSUMOylation (Figure 7b). SUMO-modified and -unmodified bands of ZFP282 were detected in the CoCoA immunoprecipitates from MCF-7 cell extracts prepared with N-ethylmaleimide, whereas a small amount of unmodified ZFP282 was precipitated by CoCoA from extracts prepared without N-ethylmaleimide. Furthermore, the ratio of SUMO-modified to unmodified ZFP282 was significantly increased in the CoCoA immunoprecipitates relative to that observed in input, suggesting that CoCoA preferentially associates with SUMOylated ZFP282. To further substantiate the SUMOylation-dependent interaction between ZFP282 and CoCoA, we performed in vitro binding assays. SUMOylated GST-ZFP282 was generated using bacterial SUMOylation system, purified, eluted and subjected to binding assays with recombinant His-tagged CoCoA (Figure 7c). SUMO-modified and unmodified ZFP282 bound CoCoA. However, the ratios of SUMO-modified to unmodified ZFP282 were significantly increased in the CoCoA-bound fraction relative to those observed in input. Interestingly, ZFP282 SUMOylation also enhanced binding to ERα (Figure 7d), suggesting that SUMOylation promotes the interaction of ZFP282 with CoCoA and ERα.
SUMOylation of ZFP282 positively regulates its interaction with CoCoA and ERα. (a) 293T cells were transfected with expression vectors as indicated. Cell extracts were prepared in the presence of N-ethylmaleimide (NEM) and immunoprecipitated with anti-ZFP282 antibody. The immunoprecipitates and inputs were analyzed by immunoblot using antibodies as indicated. (b) MCF-7 cell extracts were prepared in the presence or absence of N-ethylmaleimide as indicated. Equal amounts of extracts were immunoprecipitated with anti-CoCoA antibody, followed by immunoblot using the indicated antibodies. The arrowhead and arrow correspond to the SUMOylated and unmodified ZFP282, respectively. The intensity of SUMOylated and unmodified ZFP282 bands was quantified by densitometry, and the ratio of SUMOylated to unmodified ZFP282 band is indicated. (c, d) Pull-down assays. SUMOylated GST-ZFP282 proteins were incubated with immobilized recombinant His-CoCoA (c) or His-ERα-LBD (d). Bound ZFP282 proteins were analyzed by immunoblot with anti-ZFP282 antibody. The arrowhead and arrow correspond to the SUMOylated and unmodified ZFP282, respectively. The intensity of SUMOylated and unmodified ZFP282 bands was quantified by densitometry, and the ratio of SUMOylated to unmodified ZFP282 band is indicated. (e) 293T cells were transfected with expression vectors as indicated. Cell extracts were prepared in the presence of N-ethylmaleimide and immunoprecipitated with anti-ERα antibody. The immunoprecipitates and inputs were analyzed by immunoblot with the indicated antibodies. (f) SUMOylation of ZFP282 facilitates recruitment of ZFP282 and CoCoA to the pS2 promoter. MCF-7 cells were transfected with pFLAG.CMV4-ZFP282 WT or 3KR and treated with E2. ChIP assays using the indicated antibodies and quantitative PCR (qPCR) analyses were performed as in Figure 2d. Data are means±s.d. (n=3). Protein levels were monitored by immunoblot using the indicated antibodies. (g) The proposed model for SUMOylation-dependent regulation of ERα-ZFP282-CoCoA complex assembly. SUMOylation of ZFP282 increases its binding affinity for CoCoA and ERα, thereby contributing to stabilization of ERα-ZFP282–CoCoA complex formation on ERα target promoters. Bold arrows represent strong interactions, and dashed arrows represent weak interactions.
Because CoCoA associates indirectly with ERα through primary co-activators5 and ZFP282 (Supplementary Figure S10) and because SUMOylation of ZFP282 enhances the interaction with ERα, we further examined the SUMOylation-dependent interaction between ZFP282 and ERα and tested ZFP282 SUMOylation-dependent association between ERα and CoCoA. In CoIP experiments, the 3KR mutant exhibited reduced binding to ERα compared with that of ZFP282 WT, and the association between ERα and CoCoA was also reduced when the 3KR mutant was expressed instead of ZFP282 WT (Figure 7e). Consistent with these results, ZFP282 3KR mutant showed reduced recruitment to the pS2 promoter, and, importantly, CoCoA recruitment to the promoter was also reduced in MCF-7 cells expressing ZFP282 3KR compared with that in MCF-7 cells expressing ZFP282 WT (Figure 7f). Taken together, these results not only support the notion that the binding of ZFP282 to ERα at the pS2 promoter is SUMOylation-dependent but also suggest that ZFP282 SUMOylation has a critical role in the recruitment of CoCoA onto the pS2 promoter.
Discussion
Transcriptional regulation by ZFP282
Transcriptional activation is tightly controlled by a complex network of regulatory events, including dynamic interactions between transcription factors and co-regulators and different post-translational modifications of components of transcription complexes. Co-activators have specific domains that anchor them to the promoter through protein–protein interactions and other domains that act upon the chromatin or transcription machinery.4 CoCoA has an important role as a secondary co-activator for NRs. The central coiled-coil domain of CoCoA anchors it to the p160 co-activators, which interact with NRs, and CoCoA potentiates NR-mediated transcription by cooperating synergistically with GRIP1, p300, CCAR1 and DBC1.5, 6, 7, 8 In this study, we identified ZFP282 as a novel CoCoA-interacting protein, and our results demonstrate that ZFP282 interacts with both the C-terminal and N-terminal ADs of CoCoA (Figure 1d). We previously showed that the extreme C-terminal sequence (F-rich region) of CoCoA is critical for its transactivation function and suggested that the F-rich sequence may be important to recruit other co-regulators that are required for its strong transactivation function.6 Here, we further demonstrate that ZFP282 binds to the F-rich sequence (Supplementary Figure S1); furthermore ZFP282 enhances (Figure 2c) and is required for the strong transactivation function of CoCoA C-terminal AD (Figure 3d).
ZFP282 functions as an ERα co-activator (Figure 2). ZFP282 is recruited to the promoters of native ERα target genes and forms a complex with CoCoA on the promoters. ZFP282 enhanced the transcriptional activity of ERα and cooperated synergistically with CoCoA, thus demonstrating the functional importance of ZFP282–CoCoA interaction. Depletion of ZFP282 caused reduction in E2-induced expression of endogenous ERα target genes and attenuated hormone-dependent growth and cell cycle progression of breast cancer cells (Figures 3 and 4). Furthermore, ZFP282 silencing reduced the survival and tumorigenic potential of breast cancer cells (Figure 4). These results firmly established ZFP282 as an ERα co-activator that has an important physiological role in breast cancer cell growth and tumorigenesis.
ZFP282 was originally identified as a transcription factor that negatively regulates HTLV-I LTR-mediated transcription by binding to U5RE.9 However, in this study, we showed that although ZFP282, as a transcription factor, appears to be a weak activator, it can positively regulate U5RE-mediated transcription (Supplementary Figures S4 and S8). Although the reason for these conflicting results is currently unknown, it is possible that the effect of ZFP282 on U5RE-mediated transcription likely depends on both its weak transactivation function as well as its ability to compete for U5RE occupancy with other strong activators. Consistent with this idea, a previous report showed that ZFP282 shares with a strong activator Sp1 the ability to bind to U5RE and represses U5RE-mediated transcription in the presence of Sp1 but not in the absence of Sp1.13 Similarly, another Krüppel-type zinc finger protein ZBP-89 has been shown to compete with Sp1 for DNA binding and oppose transcriptional activation by Sp1 but also has been shown to function as a modest activator in the absence of Sp1.14 ZFP398, a paralog of ZFP282, was also shown to modestly activate U5RE-like element-driven reporter expression.15 Thus, although on its own ZFP282 appears to be a weak activator, it has the capacity to exert quite substantial effects on U5RE-mediated transcriptional activation when it is SUMOylated and complexed with CoCoA (Supplementary Figures S4 and S8).
Functional relevance of ZFP282 SUMOylation
SUMO has been shown to covalently modify a large number of proteins with important roles in transcriptional regulation, and SUMO modification is generally associated with transcriptional repression.1, 2 In many cases, part of this mechanism has been shown to be due to SUMO-dependent recruitment of co-repressors including HDACs and DAXX.16, 17 For example, HDACs and DAXX are recruited to SUMO-modified transcription factors and co-regulators in a SUMO-dependent manner and induce changes in chromatin structure associated with transcriptional repression. Although there are now a growing number of cases where SUMOylation has been shown to activate transcription, the underlying mechanisms involved remain unclear.
ZFP282 contains three SUMO consensus sites in the IR domain (Figure 1b) and undergoes SUMOylation on these conserved lysine residues (Figure 5). Importantly, we found that SUMOylation positively regulates the co-activator activity of ZFP282 (Figure 6). ERα and CoCoA preferentially bound to SUMOylated ZFP282, and SUMOylation-defective 3KR mutant exhibited less binding ability with ERα and CoCoA (Figure 7). These data clearly indicate that SUMOylation of ZFP282, at least in part, may contribute to stabilization of ERα-ZFP282–CoCoA complex formation. In agreement with this notion, the 3KR mutant was less recruited to pS2 promoter than was ZFP282 WT, thereby reducing CoCoA recruitment to the promoter. It is therefore tempting to speculate that SUMOylation induces a conformational change in ZFP282 that increases its binding affinity for ERα and CoCoA (Figure 7g). The reason why ZFP282 harbors multiple, closely spaced SUMOylation sites is unclear. Possibly, the repeated SUMOylation sites may serve as a fail-safe or gauge-like mechanism to ensure SUMOylation of ZFP282 and to dynamically control its co-activator activity. Together, our findings showing SUMOylation-dependent transcription complex formation provide an additional layer of regulation of co-activator complex assembly and function.
The SUMO pathway is hyperactivated in breast cancer.18 It is becoming increasingly clear that SUMOylation controls the activity of different proteins involved in ERα pathways in a complex manner. Overexpression of SUMO E2 Ubc9 in MCF-7 cells led to larger tumors in a xenograft mouse model, while a dominant negative Ubc9 suppressed tumor growth.19 These data suggest that a hyperactivated SUMO pathway contributes to breast tumor growth, possibly by stimulating ERα-dependent transcription that is activated by the SUMO pathway. ERα was shown to be SUMOylated in a hormone-dependent manner, and blocking its SUMOylation impaired ERα-induced transcription through decreased DNA binding.20 NR co-activators GRIP1 and CoCoA were also reported to be SUMOylated, and SUMOylation enhances their co-activator activity.12, 21 In this study, we showed that ZFP282 is required for tumorigenic potential of breast cancer cells and that SUMOylation potentiates co-activator function of ZFP282 for ERα (Figure 6). Thus, SUMOylation of ERα and its co-activators is likely to be relevant in breast tumorigenesis. However, as negative effects of SUMOylation on other ERα co-activators, such as AIB1, p300 and CBP, have been also reported16, 17, 22 and as SUMOylation and deSUMOylation exist in a dynamic equilibrium, the roles of SUMOylation in estrogen signaling, positive and negative, are likely to be very complex. Thus, further elucidation of the biological importance of co-regulator SUMOylation and their roles in cancer progression will provide useful information for understanding disease mechanisms and developing therapeutic drugs.
Materials and methods
Isolation of ZFP282
Yeast two-hybrid screening was performed according to the procedure described previously.5 The C-terminal AD fragment (amino acid 661–691) of CoCoA was fused to Gal4-DNA-binding domain in pGBT9 and used as bait. Yeast strain HF7c (containing his3 and lacZ reporters controlled by Gal4 response elements) was sequentially transformed with pGBT9-CoCoA AD and a mouse 17-day embryo cDNA library in pGAD10 (Clontech, Mountain View, CA, USA). 2 × 106 transformants were plated onto synthetic complete media plates lacking histidines, leucine and tryptophan, and containing 120 mM 3-amino-1,2,4-triazole to suppress background expression of His3 because of strong autonomous transactivation activity of the bait. ZFP282 was represented by three of the 25 His- and LacZ-positive clones and was the most frequently isolated clone.
SUMOylation assays
In vivo SUMOylation assay was performed, as described previously.12 COS-7 cells were transfected with EGFP-SUMO1 and the indicated HA-tagged ZFP282 expression plasmids. Cell extracts were prepared in the presence of 20 mM N-ethylmaleimide. Immunoblots were performed with anti-HA antibody. To generate SUMO1-modified GST-ZFP282 proteins in Escherichia coli, pGEX-5X-1-ZFP282 and pT-E1E2S1 (encoding E1, E2 and SUMO1GG) were introduced into E. coli BL21 (DE3), and cotransformants were selected on ampicillin/chloramphenicol plates. GST-ZFP282 proteins were expressed, purified and eluted, as described previously23 and analyzed by SDS–Polyacrylamide gel electrophoresis, followed by Coomassie blue staining.
Protein–protein interaction assays and immunoblot
For CoIP assays, COS-7, 293T or MCF-7 cell extracts were immunoprecipitated by specific antibodies or control IgG and protein G Dynabeads (Invitrogen, Grand Island, NY, USA), as indicated in figure legends. For GST pull-down assays, HA-tagged proteins were synthesized in vitro by using TNT-Quick-coupled transcription/translation system (Promega, San Luis Obispo, CA, USA) and incubated with immobilized GST-fusion proteins. After washing, bound proteins were analyzed by immunoblot with anti-HA antibody. For in vitro His pull-down assays, SUMO1-modified and -unmodified GST-ZFP282 proteins were incubated with recombinant His-tagged CoCoA or His-tagged ERα-LBD bound to Ni-NTA agarose beads (Qiagen, Valencia, CA, USA). After washing, bound proteins were analyzed by immunoblot with anti-ZFP282 antibody. The intensity of SUMO-modified and -unmodified ZFP282 was quantified by densitometry.
References
Geiss-Friedlander R, Melchior F . Concepts in sumoylation: a decade on. Nat Rev Mol Cell Biol 2007; 8: 947–956.
Gill G . Something about SUMO inhibits transcription. Curr Opin Genet Dev 2005; 15: 536–541.
Tsai MJ, O’Malley BW . Molecular mechanisms of action of steroid/thyroid receptor superfamily members. Annu Rev Biochem 1994; 63: 451–486.
Stallcup MR, Kim JH, Teyssier C, Lee YH, Ma H, Chen D . The roles of protein-protein interactions and protein methylation in transcriptional activation by nuclear receptors and their coactivators. J Steroid Biochem Mol Biol 2003; 85: 139–145.
Kim JH, Li H, Stallcup MR . CoCoA a nuclear receptor coactivator which acts through an N-terminal activation domain of p160 coactivators. Mol Cell 2003; 12: 1537–1549.
Kim JH, Yang CK, Stallcup MR . Downstream signaling mechanism of the C-terminal activation domain of transcriptional coactivator CoCoA. Nucleic Acids Res 2006; 34: 2736–2750.
Kim JH, Yang CK, Heo K, Roeder RG, An W, Stallcup MR . CCAR1 a key regulator of mediator complex recruitment to nuclear receptor transcription complexes. Mol Cell 2008; 31: 510–519.
Yu EJ, Kim SH, Heo K, Ou CY, Stallcup MR, Kim JH . Reciprocal roles of DBC1 and SIRT1 in regulating estrogen receptor α activity and co-activator synergy. Nucleic Acids Res 2011; 39: 6932–6943.
Okumura K, Sakaguchi G, Naito K, Tamura T, Igarashi H . HUB1 a novel Kruppel type zinc finger protein, represses the human T cell leukemia virus type I long terminal repeat-mediated expression. Nucleic Acids Res 1997; 25: 5025–5032.
Buache E, Etique N, Alpy F, Stoll I, Muckensturm M, Reina-San-Martin B et al. Deficiency in trefoil factor 1 (TFF1) increases tumorigenicity of human breast cancer cells and mammary tumor development in TFF1-knockout mice. Oncogene 2011; 30: 3261–3273.
Li S, Shang Y . Regulation of SRC family coactivators by post-translational modifications. Cell Signal 2007; 19: 1101–1112.
Yang CK, Kim JH, Ann DK, Stallcup MR . Differential regulation of the two transcriptional activation domains of the coiled-coil coactivator CoCoA by sumoylation. BMC Mol Biol 2008; 9: 12.
Tamura T, Sakata T, Igarashi H, Okumura K . Transcription factor HUB1 represses SP1-mediated gene expression through the CACCC box of HTLV-I U5RE but not the GC box. J Health Sci 2004; 50: 417–422.
Chupreta S, Brevig H, Bai L, Merchant JL, Iniguez-Lluhi JA . Sumoylation-dependent control of homotypic and heterotypic synergy by the Kruppel-type zinc finger protein ZBP-89. J Biol Chem 2007; 282: 36155–36166.
Conroy AT, Sharma M, Holtz AE, Wu C, Sun Z, Weigel RJ . A novel zinc finger transcription factor with two isoforms that are differentially repressed by estrogen receptor-alpha. J Biol Chem 2002; 277: 9326–9334.
Kuo HY, Chang CC, Jeng JC, Hu HM, Lin DY, Maul GG et al. SUMO modification negatively modulates the transcriptional activity of CREB-binding protein via the recruitment of Daxx. Proc Natl Acad Sci USA 2005; 102: 16973–16978.
Girdwood D, Bumpass D, Vaughan OA, Thain A, Anderson LA, Snowden AW et al. P300 transcriptional repression is mediated by SUMO modification. Mol Cell 2003; 11: 1043–1054.
Karamouzis MV, Konstantinopoulos PA, Badra FA, Papavassiliou AG . SUMO and estrogen receptors in breast cancer. Breast Cancer Res Treat 2008; 107: 195–210.
Mo YY, Yu Y, Theodosiou E, Ee PL, Beck WT . A role for Ubc9 in tumorigenesis. Oncogene 2005; 24: 2677–2683.
Sentis S, Le Romancer M, Bianchin C, Rostan MC, Corbo L . Sumoylation of the estrogen receptor alpha hinge region regulates its transcriptional activity. Mol Endocrinol 2005; 19: 2671–2684.
Kotaja N, Karvonen U, Janne OA, Palvimo JJ . The nuclear receptor interaction domain of GRIP1 is modulated by covalent attachment of SUMO-1. J Biol Chem 2002; 277: 30283–30288.
Wu H, Sun L, Zhang Y, Chen Y, Shi B, Li R et al. Coordinated regulation of AIB1 transcriptional activity by sumoylation and phosphorylation. J Biol Chem 2006; 281: 21848–21856.
Uchimura Y, Nakamura M, Sugasawa K, Nakao M, Saitoh H . Overproduction of eukaryotic SUMO-1- and SUMO-2-conjugated proteins in Escherichia coli. Anal Biochem 2004; 331: 204–206.
Acknowledgements
We thank Dr Geoffrey L Greene (University of Chicago) for pET-23d-hERα-LBD, Dr David K Ann (City of Hope Beckman Research Institute) for SENP1 and Ubc9 expression vectors, Dr Hisato Saitoh (Kimamoto University) for pT-E1E2S1, and Dr Ichi Fujisawa (Kansai Medical University) for pLTR-GL3. This research was supported by Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science and Technology (2010-0021428) (to JHK), by the Samsung Biomedical Research Institute grant (GE1-B2-061 to JHK), by Korea Healthcare Technology R&D Project funded by the Ministry for Health & Welfare (A092255), and by grant DK43093 from the United States National Institutes of Health (to MRS).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on the Oncogene website
Supplementary information
Rights and permissions
This work is licensed under the Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/3.0/
About this article
Cite this article
Yu, E., Kim, SH., Kim, M. et al. SUMOylation of ZFP282 potentiates its positive effect on estrogen signaling in breast tumorigenesis. Oncogene 32, 4160–4168 (2013). https://doi.org/10.1038/onc.2012.420
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/onc.2012.420
Keywords
This article is cited by
-
Neighborhood characteristics and breast tumor methylation: using epigenomics to explore cancer outcome disparities
Breast Cancer Research and Treatment (2022)
-
Tripartite motif-containing 3 (TRIM3) enhances ER signaling and confers tamoxifen resistance in breast cancer
Oncogenesis (2021)
-
DUF3669, a “domain of unknown function” within ZNF746 and ZNF777, oligomerizes and contributes to transcriptional repression
BMC Molecular and Cell Biology (2019)
-
Positive regulation of β-catenin–PROX1 signaling axis by DBC1 in colon cancer progression
Oncogene (2016)
-
A positive role of DBC1 in PEA3-mediated progression of estrogen receptor-negative breast cancer
Oncogene (2015)