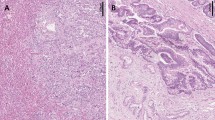
Abstract
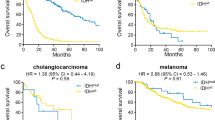
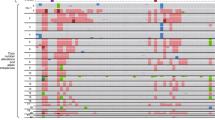
Mutations in the genes encoding isocitrate dehydrogenase, IDH1 and IDH2, have been reported in gliomas, myeloid leukemias, chondrosarcomas and thyroid cancer. We discovered IDH1 and IDH2 mutations in 34 of 326 (10%) intrahepatic cholangiocarcinomas. Tumor with mutations in IDH1 or IDH2 had lower 5-hydroxymethylcytosine and higher 5-methylcytosine levels, as well as increased dimethylation of histone H3 lysine 79 (H3K79). Mutations in IDH1 or IDH2 were associated with longer overall survival (P=0.028) and were independently associated with a longer time to tumor recurrence after intrahepatic cholangiocarcinoma resection in multivariate analysis (P=0.021). IDH1 and IDH2 mutations were significantly associated with increased levels of p53 in intrahepatic cholangiocarcinomas, but no mutations in the p53 gene were found, suggesting that mutations in IDH1 and IDH2 may cause a stress that leads to p53 activation. We identified 2309 genes that were significantly hypermethylated in 19 cholangiocarcinomas with mutations in IDH1 or IDH2, compared with cholangiocarcinomas without these mutations. Hypermethylated CpG sites were significantly enriched in CpG shores and upstream of transcription start sites, suggesting a global regulation of transcriptional potential. Half of the hypermethylated genes overlapped with DNA hypermethylation in IDH1-mutant gliobastomas, suggesting the existence of a common set of genes whose expression may be affected by mutations in IDH1 or IDH2 in different types of tumors.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Yan H, Parsons DW, Jin GL, McLendon R, Rasheed BA, Yuan WS et al. IDH1 and IDH2 mutations in gliomas. N Engl J Med 2009; 360: 765–773.
Marcucci G, Maharry K, Wu YZ, Radmacher MD, Mrozek K, Margeson D et al. IDH1 and IDH2 gene mutations identify novel molecular subsets within de novo cytogenetically normal acute myeloid leukemia: a Cancer and Leukemia Group B Study. J Clin Oncol 2010; 28: 2348–2355.
Amary MF, Bacsi K, Maggiani F, Damato S, Halai D, Berisha F et al. IDH1 and IDH2 mutations are frequent events in central chondrosarcoma and central and periosteal chondromas but not in other mesenchymal tumours. J Pathol 2011; 224: 334–343.
Shibata T, Kokubu A, Miyamoto M, Sasajima Y, Yamazaki N . Mutant IDH1 confers an in vivo growth in a melanoma cell line with BRAF mutation. Am J Pathol 2011; 178: 1395–1402.
Amary MF, Damato S, Halai D, Eskandarpour M, Berisha F, Bonar F et al. Ollier disease and Maffucci syndrome are caused by somatic mosaic mutations of IDH1 and IDH2. Nat Genet 2011; 43: 1262–1265.
Pansuriya TC, van Eijk R, d'Adamo P, van Ruler MA, Kuijjer ML, Oosting J et al. Somatic mosaic IDH1 and IDH2 mutations are associated with enchondroma and spindle cell hemangioma in Ollier disease and Maffucci syndrome. Nat Genet 2011; 43: 1256–1261.
Zhao SM, Lin Y, Xu W, Jiang WQ, Zha ZY, Wang P et al. Glioma-derived mutations in IDH1 dominantly inhibit IDH1 catalytic activity and induce HIF-1 alpha. Science 2009; 324: 261–265.
Dang L, White DW, Gross S, Bennett BD, Bittinger MA, Driggers EM et al. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature 2009; 462: 739–744.
Xu W, Yang H, Liu Y, Yang Y, Wang P, Kim SH et al. Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of alpha-ketoglutarate-dependent dioxygenases. Cancer Cell 2011; 19: 17–30.
Figueroa ME, Abdel-Wahab O, Lu C, Ward PS, Patel J, Shih A et al. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoeitic differentiation. Cancer Cell 2010; 18: 553–567.
Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science 2009; 324: 930–935.
Ito S, D'Alessio AC, Taranova OV, Hong K, Sowers LC, Zhang Y . Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature 2010; 466: 1129–1133.
Ito S, Shen L, Dai Q, Wu SC, Collins LB, Swenberg JA et al. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science 2011; 333: 1300–1303.
He YF, Li BZ, Li Z, Liu P, Wang Y, Tang Q et al. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science 2011; 333: 1303–1307.
Noushmehr H, Weisenberger DJ, Diefes K, Phillips HS, Pujara K, Berman BP et al. Identification of a CpG Island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell 2010; 17: 510–522.
Verhaak RGW, Hoadley KA, Purdom E, Wang V, Qi Y, Wilkerson MD et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell 2009; 17: 98–110.
Parsons DW, Jones S, Zhang X, Lin JC, Leary RJ, Angenendt P et al. An integrated genomic analysis of human glioblastoma multiforme. Science 2008; 321: 1807–1812.
Toyota M, Ahuja N, Ohe-Toyota M, Herman JG, Baylin SB, Issa JP . CpG island methylator phenotype in colorectal cancer. Proc Natl Acad Sci USA 1999; 96: 8681–8686.
Nagasaka T, Sasamoto H, Notohara K, Cullings HM, Takeda M, Kimura K et al. Colorectal cancer with mutation in BRAF, KRAS, and wild-type with respect to both oncogenes showing different patterns of DNA methylation. J Clin Oncol 2004; 22: 4584–4594.
Jones PA, Baylin SB . The epigenomics of cancer. Cell 2007; 128: 683–692.
Widschwendter M, Fiegl H, Egle D, Mueller-Holzner E, Spizzo G, Marth C et al. Epigenetic stem cell signature in cancer. Nature Genet 2007; 39: 157–158.
Ohm JE, McGarvey KM, Yu X, Cheng LZ, Schuebel KE, Cope L et al. A stem cell-like chromatin pattern may predispose tumor suppressor genes to DNA hypermethylation and heritable silencing. Nat Genet 2007; 39: 237–242.
Schlesinger Y, Straussman R, Keshet I, Farkash S, Hecht M, Zimmerman J et al. Polycomb-mediated methylation on Lys27 of histone H3 pre-marks genes for de novo methylation in cancer. Nat Genet 2007; 39: 232–236.
Fang F, Turcan S, Rimner A, Kaufman A, Giri D, Morris LG et al. Breast cancer methylomes establish an epigenomic foundation for metastasis. Sci Transl Med 2011; 3: 75ra25.
Blechacz B, Gores GJ . Cholangiocarcinoma: advances in pathogenesis, diagnosis, and treatment. Hepatology 2008; 48: 308–321.
Hezel AF, Deshpande V, Zhu AX . Genetics of biliary tract cancers and emerging targeted therapies. J Clin Oncol 2010; 28: 3531–3540.
Yang B, Zhong C, Peng YJ, Lai Z, Ding JP . Molecular mechanisms of ‘off-on switch’ of activities of human IDH1 by tumor-associated mutation R132H. Cell Res 2010; 20: 1188–1200.
Xu Y, Wu F, Tan L, Kong L, Xiong L, Deng J et al. Genome-wide regulation of 5hmC, 5mC, and gene expression by Tet1 hydroxylase in mouse embryonic stem cells. Mol Cell 2011; 42: 451–464.
Dunn GP, Rinne ML, Wykosky J, Genovese G, Quayle SN, Dunn IF et al. Emerging insights into the molecular and cellular basis of glioblastoma. Genes Dev 2012; 26: 756–784.
Watanabe T, Nobusawa S, Kleihues P, Ohgaki H . IDH1 mutations are early events in the development of astrocytomas and oligodendrogliomas. Am J Pathol 2009; 174: 1149–1153.
Aishima S, Kuroda Y, Nishihara Y, Taguchi K, Taketomi A, Maehara Y et al. Gastric mucin phenotype defines tumour progression and prognosis of intrahepatic cholangiocarcinoma: gastric foveolar type is associated with aggressive tumour behaviour. Histopathology 2006; 49: 35–44.
Andersen JB, Spee B, Bechacz BR, Avital I, Komuta M, Barbour A et al. Genomic and genetic characterization of cholangiocarcinoma identifies therapeutic targets for tyrosine kinase inhibitors. Gastroenterology 2012; 142: 1021–1031.
Borger DR, Tanabe KK, Fan KC, Lopez HU, Fantin VR, Straley KS et al. Frequent mutation of isocitrate dehydrogenase (IDH)1 and IDH2 in cholangiocarcinoma identified through broad-based tumor genotyping. Oncologis 2012; 17: 72–79.
Ward PS, Patel J, Wise DR, Abdel-Wahab O, Bennett BD, Coller HA et al. The common feature of leukemia-associated IDH1 and IDH2 mutations is a neomorphic enzyme activity converting alpha-ketoglutarate to 2-hydroxyglutarate. Cancer Cell 2010; 17: 225–234.
Kipp BR, Voss JS, Kerr SE, Barr Fritcher EG, Graham RP, Zhang L et al. Isocitrate dehydrogenase 1 and 2 mutations in cholangiocarcinoma. Hum Pathol 2012.
Moran-Crusio K, Reavie L, Shih A, Abdel-Wahab O, Ndiaye-Lobry D, Lobry C et al. Tet2 loss leads to increased hematopoietic stem cell self-renewal and myeloid transformation. Cancer Cell 2011; 20: 11–24.
Abdel-Wahab O, Manshouri T, Patel J, Harris K, Yao JJ, Hedvat C et al. Genetic analysis of transforming events that convert chronic myeloproliferative neoplasms to leukemias. Cancer Res 2010; 70: 447–452.
Turner R, Lozoya O, Wang Y, Cardinale V, Gaudio E, Alpini G et al. Human hepatic stem cell and maturational liver lineage biology. Hepatology 2011; 53: 1035–1045.
Cardinale V, Wang Y, Carpino G, Mendel G, Alpini G, Gaudio E et al. The biliary tree-a reservoir of multipotent stem cells. Nat Rev Gastroenterol Hepatol 2012; 9: 231–240.
Hinoue T, Weisenberger DJ, Lange CP, Shen H, Byun HM, Van Den Berg D et al. Genome-scale analysis of aberrant DNA methylation in colorectal cancer. Genome Res 2012; 22: 271–282.
Brenet F, Moh M, Funk P, Feierstein E, Viale AJ, Socci ND et al. DNA methylation of the first exon is tightly linked to transcriptional silencing. PLoS One 2011; 6: e14524.
Chapman MA, Lawrence MS, Keats JJ, Cibulskis K, Sougnez C, Schinzel AC et al. Initial genome sequencing and analysis of multiple myeloma. Nature 2011; 471: 467–472.
Llovet JM, Di Bisceglie AM, Bruix J, Kramer BS, Lencioni R, Zhu AX et al. Design and endpoints of clinical trials in hepatocellular carcinoma. J Natl Cancer Inst 2008; 100: 698–711.
Andersen JB, Factor VM, Marquardt JU, Raggi C, Lee YH, Seo D et al. An integrated genomic and epigenomic approach predicts therapeutic response to zebularine in human liver cancer. Sci Transl Med 2010; 2: 54ra77.
Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA 2005; 102: 15545–15550.
Merico D, Isserlin R, Stueker O, Emili A, Bader GD . Enrichment map: a network-based method for gene-set enrichment visualization and interpretation. PLoS One 2010; 5: e13984.
Acknowledgements
The UNC Biospecimen Core Facility, Mammalian Genotyping Core Facility and Translational Pathology Laboratory provided technical assistance for this study. Funding for this research was provided by: the 985 Program from the Chinese Ministry of Education (KLG, YX), MOST 973 (No. 2009CB918401, No. 2011CB910600, No. 2012CB910300, No. 2012CB910101; DY, YX and KLG); the NSFC Program of International Cooperation and Exchanges (No. 81120108016; LXQ, YX) and the China National Key Projects for Infectious Disease (2008ZX10002-021, 2012ZX10002-012; LZQ), the National Institutes of Health (KLG, YX, SST, LRR), the James S McDonnell Foundation (YX), the Samuel Waxman Cancer Research Foundation (YX), the American Gastroenterological Association Foundation for Digestive Health and Nutrition (LRR), the Alfred P Sloan Foundation fellowship (DYC), the Nancy Stegman Cancer Research Fund (DYC) and the University Cancer Research Fund (DYC, YX).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
DYC is a consultant for Aveo Pharmaceuticals and Entremed, and held minor stock ownership in Illumina, which did not have any role in the study design, data collection and analysis, decision to publish or preparation of the manuscript.
Additional information
Supplementary Information accompanies the paper on the Oncogene website
Supplementary information
Rights and permissions
About this article
Cite this article
Wang, P., Dong, Q., Zhang, C. et al. Mutations in isocitrate dehydrogenase 1 and 2 occur frequently in intrahepatic cholangiocarcinomas and share hypermethylation targets with glioblastomas. Oncogene 32, 3091–3100 (2013). https://doi.org/10.1038/onc.2012.315
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/onc.2012.315
Keywords
This article is cited by
-
Pharmacokinetics/pharmacodynamics of ivosidenib in advanced IDH1-mutant cholangiocarcinoma: findings from the phase III ClarIDHy study
Cancer Chemotherapy and Pharmacology (2024)
-
The role and implication of autophagy in cholangiocarcinoma
Cell Death Discovery (2023)
-
Impact of IDH1 mutation on clinical course of patients with intrahepatic cholangiocarcinoma: a retrospective analysis from a German tertiary center
Journal of Cancer Research and Clinical Oncology (2023)
-
Clinical Outcomes After Progression on First-Line Therapies in IDH1 Mutated Versus Wild-Type Intrahepatic Cholangiocarcinoma Patients
Targeted Oncology (2023)
-
Significant Response to Camrelizumab Plus Targeted Drugs in Recurrent Intrahepatic Cholangiocarcinoma: a Case Report and Literature Review
Journal of Gastrointestinal Cancer (2022)