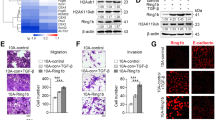
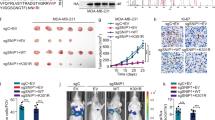
Abstract
Expression of E-cadherin, a hallmark of epithelial–mesenchymal transition (EMT), is often lost due to promoter DNA methylation in basal-like breast cancer (BLBC), which contributes to the metastatic advantage of this disease; however, the underlying mechanism remains unclear. Here, we identified that Snail interacted with Suv39H1 (suppressor of variegation 3-9 homolog 1), a major methyltransferase responsible for H3K9me3 that intimately links to DNA methylation. We demonstrated that the SNAG domain of Snail and the SET domain of Suv39H1 were required for their mutual interactions. We found that H3K9me3 and DNA methylation on the E-cadherin promoter were higher in BLBC cell lines. We showed that Snail interacted with Suv39H1 and recruited it to the E-cadherin promoter for transcriptional repression. Knockdown of Suv39H1 restored E-cadherin expression by blocking H3K9me3 and DNA methylation and resulted in the inhibition of cell migration, invasion and metastasis of BLBC. Our study not only reveals a critical mechanism underlying the epigenetic regulation of EMT, but also paves a way for the development of new treatment strategies against this disease.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Abbreviations
- BLBC:
-
basal-like breast cancer
- ChIP:
-
chromatin immunoprecipitation
- EMT:
-
epithelial–mesenchymal transition
- LSD1:
-
lysine-specific demethylase 1
- Suv39H1:
-
suppressor of variegation 3-9 homolog 1.
References
Jemal A, Siegel R, Ward E, Hao Y, Xu J, Murray T et al. Cancer statistics, 2008. CA Cancer J Clin 2008; 58: 71–96.
Nieto MA . The snail superfamily of zinc-finger transcription factors. Nat Rev Mol Cell Biol 2002; 3: 155–166.
Peinado H, Olmeda D, Cano A . Snail, Zeb and bHLH factors in tumour progression: an alliance against the epithelial phenotype? Nat Rev Cancer 2007; 7: 415–428.
Thiery JP, Sleeman JP . Complex networks orchestrate epithelial-mesenchymal transitions. Nat Rev Mol Cell Biol 2006; 7: 131–142.
Kalluri R, Weinberg RA . The basics of epithelial-mesenchymal transition. J Clin Invest 2009; 119: 1420–1428.
Thiery JP, Acloque H, Huang RY, Nieto MA . Epithelial-mesenchymal transitions in development and disease. Cell 2009; 139: 871–890.
Wu Y, Zhou BP . New insights of epithelial-mesenchymal transition in cancer metastasis. Acta Biochim Biophys Sin (Shanghai) 2008; 40: 643–650.
Li X, Lewis MT, Huang J, Gutierrez C, Osborne CK, Wu MF et al. Intrinsic resistance of tumorigenic breast cancer cells to chemotherapy. J Natl Cancer Inst 2008; 100: 672–679.
Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan A, Zhou AY et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell 2008; 133: 704–715.
Moody SE, Perez D, Pan TC, Sarkisian CJ, Portocarrero CP, Sterner CJ et al. The transcriptional repressor Snail promotes mammary tumor recurrence. Cancer Cell 2005; 8: 197–209.
Polyak K, Weinberg RA . Transitions between epithelial and mesenchymal states: acquisition of malignant and stem cell traits. Nat Rev Cancer 2009; 9: 265–273.
Yang J, Weinberg RA . Epithelial-mesenchymal transition: at the crossroads of development and tumor metastasis. Dev Cell 2008; 14: 818–829.
Julien S, Puig I, Caretti E, Bonaventure J, Nelles L, van Roy F et al. Activation of NF-kappaB by Akt upregulates Snail expression and induces epithelium mesenchyme transition. Oncogene 2007; 26: 7445–7456.
Lopez-Novoa JM, Nieto MA . Inflammation and EMT: an alliance towards organ fibrosis and cancer progression. EMBO Mol Med 2009; 1: 303–314.
Wu Y, Deng J, Rychahou PG, Qiu S, Evers BM, Zhou BP . Stabilization of snail by NF-kappaB is required for inflammation-induced cell migration and invasion. Cancer Cell 2009; 15: 416–428.
Feinberg AP . Phenotypic plasticity and the epigenetics of human disease. Nature 2007; 447: 433–440.
Jones PA, Baylin SB . The epigenomics of cancer. Cell 2007; 128: 683–692.
Cheng X, Blumenthal RM . Coordinated chromatin control: structural and functional linkage of DNA and histone methylation. Biochemistry 2010; 49: 2999–3008.
Eissenberg JC, Shilatifard A . Histone H3 lysine 4 (H3K4) methylation in development and differentiation. Dev Biol 2010; 339: 240–249.
Laurent L, Wong E, Li G, Huynh T, Tsirigos A, Ong CT et al. Dynamic changes in the human methylome during differentiation. Genome Res 2010; 20: 320–331.
Ruthenburg AJ, Allis CD, Wysocka J . Methylation of lysine 4 on histone H3: intricacy of writing and reading a single epigenetic mark. Mol Cell 2007; 25: 15–30.
Cedar H, Bergman Y . Linking DNA methylation and histone modification: patterns and paradigms. Nat Rev Genet 2009; 10: 295–304.
McCabe MT, Brandes JC, Vertino PM . Cancer DNA methylation: molecular mechanisms and clinical implications. Clin Cancer Res 2009; 15: 3927–3937.
Bergamaschi A, Hjortland GO, Triulzi T, Sorlie T, Johnsen H, Ree AH et al. Molecular profiling and characterization of luminal-like and basal-like in vivo breast cancer xenograft models. Mol Oncol 2009; 3: 469–482.
Blick T, Widodo E, Hugo H, Waltham M, Lenburg ME, Neve RM et al. Epithelial mesenchymal transition traits in human breast cancer cell lines. Clin Exp Metastasis 2008; 25: 629–642.
Hennessy BT, Gonzalez-Angulo AM, Stemke-Hale K, Gilcrease MZ, Krishnamurthy S, Lee JS et al. Characterization of a naturally occurring breast cancer subset enriched in epithelial-to-mesenchymal transition and stem cell characteristics. Cancer Res 2009; 69: 4116–4124.
Sarrio D, Rodriguez-Pinilla SM, Hardisson D, Cano A, Moreno-Bueno G, Palacios J . Epithelial-mesenchymal transition in breast cancer relates to the basal-like phenotype. Cancer Res 2008; 68: 989–997.
Storci G, Sansone P, Trere D, Tavolari S, Taffurelli M, Ceccarelli C et al. The basal-like breast carcinoma phenotype is regulated by SLUG gene expression. J Pathol 2008; 214: 25–37.
Wang Y, Zhou BP . Epithelial-mesenchymal transition in breast cancer progression and metastasis. Chin J Cancer 2011; 30: 603–611.
Lin Y, Wu Y, Li J, Dong C, Ye X, Chi YI et al. The SNAG domain of Snail1 functions as a molecular hook for recruiting lysine-specific demethylase 1. EMBO J. 2010; 29: 1803–1816.
Rudolph T, Yonezawa M, Lein S, Heidrich K, Kubicek S, Schafer C et al. Heterochromatin formation in Drosophila is initiated through active removal of H3K4 methylation by the LSD1 homolog SU(VAR)3-3. Mol Cell 2007; 26: 103–115.
Barrallo-Gimeno A, Nieto MA . Evolutionary history of the Snail/Scratch superfamily. Trends Genet 2009; 25: 248–252.
Chin K, DeVries S, Fridlyand J, Spellman PT, Roydasgupta R, Kuo WL et al. Genomic and transcriptional aberrations linked to breast cancer pathophysiologies. Cancer Cell 2006; 10: 529–541.
Mullins M, Perreard L, Quackenbush JF, Gauthier N, Bayer S, Ellis M et al. Agreement in breast cancer classification between microarray and quantitative reverse transcription PCR from fresh-frozen and formalin-fixed, paraffin-embedded tissues. Clin Chem 2007; 53: 1273–1279.
Dillon SC, Zhang X, Trievel RC, Cheng X . The ET-domain protein superfamily: protein lysine methyltransferases. Genome Biol. 2005; 6: 227.
Shinkai Y, Tachibana M . H3K9 methyltransferase G9a and the related molecule GLP. Genes Dev 2011; 25: 781–788.
Wu H, Min J, Lunin VV, Antoshenko T, Dombrovski L, Zeng H et al. Structural biology of human H3K9 methyltransferases. PLoS One 2010; 5: e8570.
Bindels S, Mestdagt M, Vandewalle C, Jacobs N, Volders L, Noel A et al. Regulation of vimentin by SIP1 in human epithelial breast tumor cells. Oncogene 2006; 25: 4975–4985.
Xie L, Law BK, Aakre ME, Edgerton M, Shyr Y, Bhowmick NA et al. Transforming growth factor beta-regulated gene expression in a mouse mammary gland epithelial cell line. Breast Cancer Res 2003; 5: R187–R198.
Yang J, Mani SA, Donaher JL, Ramaswamy S, Itzykson RA, Come C et al. Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell 2004; 117: 927–939.
Lombaerts M, van Wezel T, Philippo K, Dierssen JW, Zimmerman RM, Oosting J et al. E-cadherin transcriptional downregulation by promoter methylation but not mutation is related to epithelial-to-mesenchymal transition in breast cancer cell lines. Br J Cancer 2006; 94: 661–671.
Kreike B, van Kouwenhove M, Horlings H, Weigelt B, Peterse H, Bartelink H et al. Gene expression profiling and histopathological characterization of triple-negative/basal-like breast carcinomas. Breast Cancer Res 2007; 9: R65.
Neve RM, Chin K, Fridlyand J, Yeh J, Baehner FL, Fevr T et al. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell 2006; 10: 515–527.
Prat A, Parker JS, Karginova O, Fan C, Livasy C, Herschkowitz JI et al. Phenotypic and molecular characterization of the claudin-low intrinsic subtype of breast cancer. Breast Cancer Res 2010; 12: R68.
Greiner D, Bonaldi T, Eskeland R, Roemer E, Imhof A . Identification of a specific inhibitor of the histone methyltransferase SU(VAR)3-9. Nat Chem Biol 2005; 1: 143–145.
Forneris F, Binda C, Battaglioli E, Mattevi A . LSD1: oxidative chemistry for multifaceted functions in chromatin regulation. Trends Biochem Sci 2008; 33: 181–189.
Martin C, Zhang Y . The diverse functions of histone lysine methylation. Nat Rev Mol Cell Biol 2005; 6: 838–849.
Dodge JE, Kang YK, Beppu H, Lei H, Li E . Histone H3-K9 methyltransferase ESET is essential for early development. Mol Cell Biol 2004; 24: 2478–2486.
Tachibana M, Sugimoto K, Nozaki M, Ueda J, Ohta T, Ohki M et al. G9a histone methyltransferase plays a dominant role in euchromatic histone H3 lysine 9 methylation and is essential for early embryogenesis. Genes Dev 2002; 16: 1779–1791.
Tachibana M, Ueda J, Fukuda M, Takeda N, Ohta T, Iwanari H et al. Histone methyltransferases G9a and GLP form heteromeric complexes and are both crucial for methylation of euchromatin at H3-K9. Genes Dev 2005; 19: 815–826.
Peters AH, Kubicek S, Mechtler K, O'Sullivan RJ, Derijck AA, Perez-Burgos L et al. Partitioning and plasticity of repressive histone methylation states in mammalian chromatin. Mol Cell 2003; 12: 1577–1589.
Rea S, Eisenhaber F, O'Carroll D, Strahl BD, Sun ZW, Schmid M et al. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature 2000; 406: 593–599.
Fritsch L, Robin P, Mathieu JR, Souidi M, Hinaux H, Rougeulle C et al. A subset of the histone H3 lysine 9 methyltransferases Suv39h1, G9a, GLP, and SETDB1 participate in a multimeric complex. Mol Cell 2010; 37: 46–56.
Kouzarides T . Chromatin modifications and their function. Cell 2007; 128: 693–705.
Fuks F . DNA methylation and histone modifications: teaming up to silence genes. Curr Opin Genet Dev 2005; 15: 490–495.
Jiao Y, Shi C, Edil BH, de Wilde RF, Klimstra DS, Maitra A et al. DAXX/ATRX, MEN1, and mTOR pathway genes are frequently altered in pancreatic neuroendocrine tumors. Science 2011; 331: 1199–1203.
Jones S, Wang TL, Shih Ie M, Mao TL, Nakayama K, Roden R et al. Frequent mutations of chromatin remodeling gene ARID1A in ovarian clear cell carcinoma. Science 2010; 330: 228–231.
Wiegand KC, Shah SP, Al-Agha OM, Zhao Y, Tse K, Zeng T et al. ARID1A mutations in endometriosis-associated ovarian carcinomas. N Engl J Med 2010; 363: 1532–1543.
Margueron R, Reinberg D . The Polycomb complex PRC2 and its mark in life. Nature 2011; 469: 343–349.
Wu Y, Evers BM, Zhou BP . Small C-terminal domain phosphatase enhances snail activity through dephosphorylation. J Biol Chem 2009; 284: 640–648.
Zhou BP, Deng J, Xia W, Xu J, Li YM, Gunduz M et al. Dual regulation of Snail by GSK-3beta-mediated phosphorylation in control of epithelial-mesenchymal transition. Nat Cell Biol 2004; 6: 931–940.
Xia W, Chen JS, Zhou X, Sun PR, Lee DF, Liao Y et al. Phosphorylation/cytoplasmic localization of p21Cip1/WAF1 is associated with HER2/neu overexpression and provides a novel combination predictor for poor prognosis in breast cancer patients. Clin Cancer Res 2004; 10: 3815–3824.
Herman JG, Graff JR, Myohanen S, Nelkin BD, Baylin SB . Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci USA 1996; 93: 9821–9826.
Eswar N, Webb B, Marti-Renom MA, Madhusudhan MS, Eramian D, Shen MY et al. Comparative protein structure modeling using Modeller. Curr Protoc Bioinformatics 2006, Chapter 5:Unit 5.6.
DeLano WL . Unraveling hot spots in binding interfaces: progress and challenges. Curr Opin Struct Biol 2002; 12: 14–20.
Acknowledgements
We thank Dr Nathan L Vanderford for critical reading and editing of this manuscript. This work was supported by grants from NIH (RO1CA125454), Susan G Komen Foundation (KG081310), and Mary Kay Ash Foundation (to BP Zhou).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Rights and permissions
About this article
Cite this article
Dong, C., Wu, Y., Wang, Y. et al. Interaction with Suv39H1 is critical for Snail-mediated E-cadherin repression in breast cancer. Oncogene 32, 1351–1362 (2013). https://doi.org/10.1038/onc.2012.169
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/onc.2012.169
Keywords
This article is cited by
-
Ubiquitylation of RUNX3 by RNA-binding ubiquitin ligase MEX3C promotes tumorigenesis in lung adenocarcinoma
Journal of Translational Medicine (2024)
-
The Wnt-pathway corepressor TLE3 interacts with the histone methyltransferase KMT1A to inhibit differentiation in Rhabdomyosarcoma
Oncogene (2024)
-
GATA zinc finger protein p66β promotes breast cancer cell migration by acting as a co-activator of Snail
Cell Death & Disease (2023)
-
Epigenetic regulation of hybrid epithelial-mesenchymal cell states in cancer
Oncogene (2023)
-
Epigenetic regulation of breast cancer metastasis
Cancer and Metastasis Reviews (2023)