Abstract
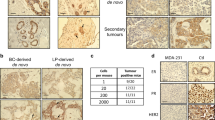
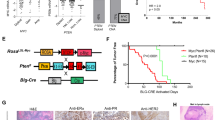
A hallmark of human cancer is heterogeneity, reflecting the complex series of changes resulting in the activation of oncogenes coupled with inactivation of tumor suppressor genes. Breast cancer is no exception and indeed, many studies have revealed considerable complexity and heterogeneity in the population of primary breast tumors and substantial changes in a recurrent breast tumor that has acquired metastatic properties and drug resistance. We have made use of a Myc-inducible transgenic mouse model of breast cancer in which elimination of Myc activity following tumor development initially leads to a regression of a subset of tumors generally followed by de novo Myc-independent growth. We have observed that tumors that grow independent of Myc expression have gene profiles that are distinct from the primary tumors with characteristics indicative of an epithelial–mesenchymal transition (EMT) phenotype. Phenotypic analyses of Myc-independent tumors confirm the acquisition of an EMT phenotype suggested to be associated with invasive and migratory properties in human cancer cells. Further genomic analyses reveal mouse mammary tumors growing independent of myc have a higher probability of exhibiting a gene signature similar to that observed for human ‘tumor-initiating’ cells. Collectively, the data reveal genetic alterations that underlie tumor progression and an escape from Myc-dependent growth in a transgenic mouse model that can provide insights to what occurs in human cancers as they acquire drug resistance and metastatic properties.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF . (2003). Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci USA 100: 3983–3988.
Andrechek ER, Cardiff RD, Chang JT, Gatza ML, Acharya CR, Potti A et al. (2009). Genetic heterogeneity of Myc-induced mammary tumors reflecting diverse phenotypes including metastatic potential. Proc Natl Acad Sci USA 106: 16387–16392.
Bild AH, Yao G, Chang JT, Wang Q, Potti A, Chasse D et al. (2006). Oncogenic pathway signatures in human cancers as a guide to targeted therapies. Nature 439: 353–357.
Boxer RB, Jang JW, Sintasath L, Chodosh LA . (2004). Lack of sustained regression of c-MYC-induced mammary adenocarcinomas following brief or prolonged MYC inactivation. Cancer Cell 6: 577–586.
Campbell LL, Polyak K . (2007). Breast tumor heterogeneity: cancer stem cells or clonal evolution? Cell Cycle 6: 2332–2338.
Cardiff RD . (2010). The pathology of EMT in mouse mammary tumorigenesis. J Mammary Gland Biol Neoplasia 15: 225–233.
Carvalho CM, Chang J, Lucas JE, Nevins JR, Wang Q, West M . (2008). High-dimensional sparse factor modeling: applications in gene expression genomics. J Am Stat Assoc 103: 1438–1456.
Creighton CJ, Li X, Landis M, Dixon JM, Neumeister VM, Sjolund A et al. (2009). Residual breast cancers after conventional therapy display mesenchymal as well as tumor-initiating features. Proc Natl Acad Sci USA 106: 13820–13825.
D'Cruz CM, Gunther EJ, Boxer RB, Hartman JL, Sintasath L, Moody SE et al. (2001). c-MYC induces mammary tumorigenesis by means of a preferred pathway involving spontaneous Kras2 mutations. Nat Med 7: 235–239.
Damonte P, Gregg JP, Borowsky AD, Keister BA, Cardiff RD . (2007). EMT tumorigenesis in the mouse mammary gland. Lab Invest 87: 1218–1226.
Diehn M, Cho RW, Clarke MF . (2009). Therapeutic implications of the cancer stem cell hypothesis. Semin Radiat Oncol 19: 78–86.
Ding L, Ellis MJ, Li S, Larson DE, Chen K, Wallis JW et al. (2010). Genome remodelling in a basal-like breast cancer metastasis and xenograft. Nature 464: 999–1005.
Ding L, Getz G, Wheeler DA, Mardis ER, McLellan MD, Cibulskis K et al. (2008). Somatic mutations affect key pathways in lung adenocarcinoma. Nature 455: 1069–1075.
Gatza ML, Lucas JE, Barry WT, Kim JW, Wang Q, Crawford MD et al. (2010). A pathway-based classification of human breast cancer. Proc Natl Acad Sci USA 107: 6994–6999.
Herschkowitz JI, Simin K, Weigman VJ, Mikaelian I, Usary J, Hu Z et al. (2007). Identification of conserved gene expression features between murine mammary carcinoma models and human breast tumors. Genome Biol 8: R76.
Huang E, Ishida S, Pittman J, Dressman H, Bild A, Kloos M et al. (2003). Gene expression phenotypic models that predict the activity of oncogenic pathways. Nat Genet 34: 226–230.
Iseri O, Kars MD, Arpaci F, Atalay C, Pak I, Gunduz U . (2011). Drug resistant MCF-7 cells exhibit epithelial-mesenchymal transition gene expression pattern. Biomed Pharmacother 65: 40–45.
Jain M, Arvanitis C, Chu K, Dewey W, Leonhardt E, Trinh M et al. (2002). Sustained loss of a neoplastic phenotype by brief inactivation of MYC. Science 297: 102–104.
Lucas J, Carvalho C, West M . (2009). A bayesian analysis strategy for cross-study translation of gene expression biomarkers. Stat Appl Genet Mol Biol 8: 1–26 (Article 11).
Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan A, Zhou AY et al. (2008). The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell 133: 704–715.
Moody SE, Perez D, Pan TC, Sarkisian CJ, Portocarrero CP, Sterner CJ et al. (2005). The transcriptional repressor Snail promotes mammary tumor recurrence. Cancer Cell 8: 197–209.
Mullighan CG, Goorha S, Radtke I, Miller CB, Coustan-Smith E, Dalton JD et al. (2007). Genome-wide analysis of genetic alterations in acute lymphoblastic leukaemia. Nature 446: 758–764.
Nicolini A, Ferrari P, Fini M, Borsari V, Fallahi P, Antonelli A et al. (2010). Stem cells: their role in breast cancer development and resistance to treatment. Curr Pharml Biotechnol 12: 196–205.
Pelengaris S, Khan M, Evan GI . (2002). Suppression of Myc-induced apoptosis in beta cells exposes multiple oncogenic properties of Myc and triggers carcinogenic progression. Cell 109: 321–334.
Podsypanina K, Politi K, Beverly LJ, Varmus HE . (2008). Oncogene cooperation in tumor maintenance and tumor recurrence in mouse mammary tumors induced by Myc and mutant Kras. Proc Natl Acad Sci USA 105: 5242–5247.
Radaelli E, Damonte P, Cardiff RD . (2009). Epithelial-mesenchymal transition in mouse mammary tumorigenesis. Future Oncol 5: 1113–1127.
Shachaf CM, Kopelman AM, Arvanitis C, Karlsson A, Beer S, Mandl S et al. (2004). MYC inactivation uncovers pluripotent differentiation and tumour dormancy in hepatocellular cancer. Nature 431: 1112–1117.
Sjoblom T, Jones S, Wood LD, Parsons DW, Lin J, Barber TD et al. (2006). The consensus coding sequences of human breast and colorectal cancers. Science 314: 268–274.
Thiery JP . (2002). Epithelial-mesenchymal transitions in tumour progression. Nat Rev Cancer 2: 442–454.
Thiery JP, Acloque H, Huang RY, Nieto MA . (2009). Epithelial-mesenchymal transitions in development and disease. Cell 139: 871–890.
Weir BA, Woo MS, Getz G, Perner S, Ding L, Beroukhim R et al. (2007). Characterizing the cancer genome in lung adenocarcinoma. Nature 450: 893–898.
Acknowledgements
The MTB/TOM mouse was a kind gift from Lewis A Chodosh. We thank Kenichiro Fujiwara for his assistance in animal husbandry and Rachel E Rempel for her helpful advice. We are grateful to Kaye Culler for her assistance with the manuscript. This work was supported by grants from the NIH (CA104663, CA112952) and grants U01 CA141541 and U01 CA141582 from the National Cancer Institute's Mouse Models of Human Cancers Consortium (RDC). We also thank Ms Katie Bell and Judith Walls of the CCM MMPL for their excellent immunohistochemistry. This work was also supported by grants from the JRN (RO1-CA104663 and U54-CA112952) from the National Cancer Institute. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Cancer Institute or the National Institutes of Health.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
Dr Nevins’ work has been funded by the NIH. He has received compensation as a member of the scientific advisory boards of the Millennium Pharmaceutical and Qiagen Scientific. In addition, he reports ownership interest in the Expression Analysis Inc. Drs Leung Andrechek and Cardiff declare no potential conflict of interest.
Additional information
Supplementary Information accompanies the paper on the Oncogene website
Supplementary information
Rights and permissions
About this article
Cite this article
Leung, J., Andrechek, E., Cardiff, R. et al. Heterogeneity in MYC-induced mammary tumors contributes to escape from oncogene dependence. Oncogene 31, 2545–2554 (2012). https://doi.org/10.1038/onc.2011.433
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/onc.2011.433
Keywords
This article is cited by
-
Clinical and molecular relevance of mutant-allele tumor heterogeneity in breast cancer
Breast Cancer Research and Treatment (2017)
-
Targeting RNA polymerase I to treat MYC-driven cancer
Oncogene (2015)
-
A genomic analysis of mouse models of breast cancer reveals molecular features ofmouse models and relationships to human breast cancer
Breast Cancer Research (2014)