Abstract
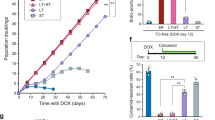
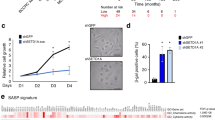
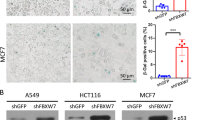
The DREAM complex is an important regulator of mitotic gene expression during the cell cycle. Here we report that inactivation of LIN9, a subunit of DREAM, results in premature senescence, which can be overcome by the SV40 large T (LT) antigen. Together with the observation that p16INK4a and p21Waf1 are upregulated upon loss of LIN9, these results indicate that senescence is triggered by the pRB and p53 tumor suppressor pathways. We also find that LIN9-null cells that escape senescence are chromosomally instable because of compromised mitotic fidelity. SV40 LT-expressing cells that adapt to the loss of LIN9 can grow anchorage-independently in soft agar, a hallmark of oncogenic transformation. Taken together, these results suggest an important role of mitotic gene regulation in the maintenance of genomic stability and tumor suppression.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Adams PD . (2009). Healing and hurting: molecular mechanisms, functions, and pathologies of cellular senescence. Mol Cell 36: 2–14.
Baker DJ, Jeganathan KB, Cameron JD, Thompson M, Juneja S, Kopecka A et al. (2004). BubR1 insufficiency causes early onset of aging-associated phenotypes and infertility in mice. Nat Genet 36: 744–749.
Blais A, Dynlacht BD . (2007). E2F-associated chromatin modifiers and cell cycle control. Curr Opin Cell Biol 19: 658–662.
Chao HH, Buchmann AM, DeCaprio JA . (2000). Loss of p19(ARF) eliminates the requirement for the pRB-binding motif in simian virus 40 large T antigen-mediated transformation. Mol Cell Biol 20: 7624–7633.
Cimini D, Degrassi F . (2005). Aneuploidy: a matter of bad connections. Trends Cell Biol 15: 442–451.
Cotsiki M, Lock RL, Cheng Y, Williams GL, Zhao J, Perera D et al. (2004). Simian virus 40 large T antigen targets the spindle assembly checkpoint protein Bub1. Proc Natl Acad Sci USA 101: 947–952.
Dimri GP, Lee X, Basile G, Acosta M, Scott G, Roskelley C et al. (1995). A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc Natl Acad Sci USA 92: 9363–9367.
Gagrica S, Hauser S, Kolfschoten I, Osterloh L, Agami R, Gaubatz S . (2004). Inhibition of oncogenic transformation by mammalian Lin-9, a pRB-associated protein. EMBO J 23: 4627–4638.
Gaubatz S, Lindeman GJ, Ishida S, Jakoi L, Nevins JR, Livingston DM et al. (2000). E2F4 and E2F5 play an essential role in pocket protein-mediated G1 control. Mol Cell 6: 729–735.
Holland AJ, Cleveland DW . (2009). Boveri revisited: chromosomal instability, aneuploidy and tumorigenesis. Nat Rev Mol Cell Biol 10: 478–487.
Kierstead TD, Tevethia MJ . (1993). Association of p53 binding and immortalization of primary C57BL/6 mouse embryo fibroblasts by using simian virus 40 T-antigen mutants bearing internal overlapping deletion mutations. J Virol 67: 1817–1829.
Knight AS, Notaridou M, Watson RJ . (2009). A Lin-9 complex is recruited by B-Myb to activate transcription of G(2)/M genes in undifferentiated embryonal carcinoma cells. Oncogene 28: 1737–1747.
Laoukili J, Kooistra MR, Bras A, Kauw J, Kerkhoven RM, Morrison A et al. (2005). FoxM1 is required for execution of the mitotic programme and chromosome stability. Nat Cell Biol 7: 126–136.
Litovchick L, Sadasivam S, Florens L, Zhu X, Swanson SK, Velmurugan S et al. (2007). Evolutionarily conserved multisubunit RBL2/p130 and E2F4 protein complex represses human cell cycle-dependent genes in quiescence. Mol Cell 26: 539–551.
Maehara K, Takahashi K, Saitoh S . (2010). CENP-A reduction induces a p53-dependent cellular senescence response to protect cells from executing defective mitoses. Mol Cell Biol 30: 2090–2104.
Musacchio A, Salmon ED . (2007). The spindle-assembly checkpoint in space and time. Nat Rev Mol Cell Biol 8: 379–393.
Osterloh L, von Eyss B, Schmit F, Rein L, Hubner D, Samans B et al. (2007). The human synMuv-like protein LIN-9 is required for transcription of G2/M genes and for entry into mitosis. EMBO J 26: 144–157.
Pilkinton M, Sandoval R, Colamonici OR . (2007a). Mammalian Mip/LIN-9 interacts with either the p107, p130/E2F4 repressor complex or B-Myb in a cell cycle-phase-dependent context distinct from the Drosophila dREAM complex. Oncogene 26: 7535–7543.
Pilkinton M, Sandoval R, Song J, Ness SA, Colamonici OR . (2007b). Mip/LIN-9 regulates the expression of B-Myb and the induction of cyclin A, cyclin B, and CDK1. J Biol Chem 282: 168–175.
Reichert N, Wurster S, Ulrich T, Schmitt K, Hauser S, Probst L et al. (2010). Lin9, a subunit of the mammalian DREAM complex, is essential for embryonic development, for survival of adult mice, and for tumor suppression. Mol Cell Biol 30: 2896–2908.
Schliekelman M, Cowley DO, O′Quinn R, Oliver TG, Lu L, Salmon ED et al. (2009). Impaired Bub1 function in vivo compromises tension-dependent checkpoint function leading to aneuploidy and tumorigenesis. Cancer Res 69: 45–54.
Schmit F, Korenjak M, Mannefeld M, Schmitt K, Franke C, Eyss BV et al. (2007). LINC, a human complex that is related to pRB-containing complexes in invertebrates regulates the expression of G2/M genes. Cell Cycle 6: 1903–1913.
Serrano M . (1997). The tumor suppressor protein p16INK4a. Exp Cell Res 237: 7–13.
Song J, Sandoval R, Pilkinton MA, Tian X, Raychaudhuri P, Colamonici OR . (2010). ARF-induced downregulation of Mip130/LIN-9 protein levels mediates a positive feedback that leads to increased expression of p16Ink4a and p19Arf. Oncogene 29: 1976–1986.
Stubdal H, Zalvide J, Campbell KS, Schweitzer C, Roberts TM, DeCaprio JA . (1997). Inactivation of pRB-related proteins p130 and p107 mediated by the J domain of simian virus 40 large T antigen. Mol Cell Biol 17: 4979–4990.
Stubdal H, Zalvide J, DeCaprio JA . (1996). Simian virus 40 large T antigen alters the phosphorylation state of the RB-related proteins p130 and p107. J Virol 70: 2781–2788.
Ye X, Zerlanko B, Zhang R, Somaiah N, Lipinski M, Salomoni P et al. (2007). Definition of pRB- and p53-dependent and -independent steps in HIRA/ASF1a-mediated formation of senescence-associated heterochromatin foci. Mol Cell Biol 27: 2452–2465.
Zalvide J, DeCaprio JA . (1995). Role of pRb-related proteins in simian virus 40 large-T-antigen-mediated transformation. Mol Cell Biol 15: 5800–5810.
Acknowledgements
We thank all members of the laboratory for their suggestions and critical reading of the manuscript. We also thank Michael Schmid and Claus Steinlein for their advice and help with karyotype analysis. This work was supported by grants from the DFG (575/6-1 and TR17-B1) to SG.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Rights and permissions
About this article
Cite this article
Hauser, S., Ulrich, T., Wurster, S. et al. Loss of LIN9, a member of the DREAM complex, cooperates with SV40 large T antigen to induce genomic instability and anchorage-independent growth. Oncogene 31, 1859–1868 (2012). https://doi.org/10.1038/onc.2011.364
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/onc.2011.364
Keywords
This article is cited by
-
LIN9 confers paclitaxel resistance in triple negative breast cancer cells by upregulating CCSAP
Science China Life Sciences (2020)
-
Human papilloma virus E7 oncoprotein abrogates the p53-p21-DREAM pathway
Scientific Reports (2017)