Abstract
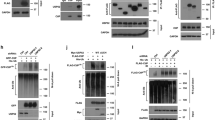
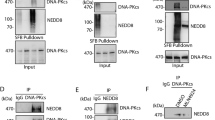
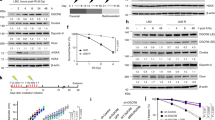
Although post-translational modifications by the small ubiquitin-like modifiers (SUMO) are known to be important in DNA damage response, it is unclear whether they have a role in double-strand break (DSB) repair by non-homologous end joining (NHEJ). Here, we analyzed various DSB repair pathways upon inhibition of SUMO-mediated protein–protein interactions using peptides that contain the SUMO-interaction motif (SIM) and discriminate between mono- and SUMO-chain modifications. The SIM peptides specifically inhibit NHEJ as shown by in vivo repair assays and radio-sensitivity of cell lines deficient in different DSB repair pathways. Furthermore, mono-SUMO, instead of SUMO-chain, modifications appear to be involved in NHEJ. Immunoprecipitation experiments also showed that the SIM peptide interacted with SUMOylated Ku70 after radiation. This study is the first to show an important role for SUMO:SIM-mediated protein–protein interactions in NHEJ, and provides a mechanistic basis for the role of SIM peptide in sensitizing genotoxic stress of cancer cells.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Andrews EA, Palecek J, Sergeant J, Taylor E, Lehmann AR, Watts FZ . (2005). Nse2, a component of the Smc5-6 complex, is a SUMO ligase required for the response to DNA damage. Mol Cell Biol 25: 185–196.
Bennardo N, Cheng A, Huang N, Stark JM . (2008). Alternative-NHEJ is a mechanistically distinct pathway of mammalian chromosome break repair. PLoS Genet 4: e1000110.
Bird AW, Yu DY, Pray-Grant MG, Qiu Q, Harmon KE, Megee PC et al. (2002). Acetylation of histone H4 by Esa1 is required for DNA double-strand break repair. Nature 419: 411–415.
Carson JP, Zhang N, Frampton GM, Gerry NP, Lenburg ME, Christman MF . (2004). Pharmacogenomic identification of targets for adjuvant therapy with the topoisomerase poison camptothecin. Cancer Res 64: 2096–2104.
Franken NA, Rodermond HM, Stap J, Haveman J, van Bree C . (2006). Clonogenic assay of cells in vitro. Nat Protoc 1: 2315–2319.
Hardeland U, Steinacher R, Jiricny J, Schar P . (2002). Modification of the human thymine-DNA glycosylase by ubiquitin-like proteins facilitates enzymatic turnover. EMBO J 21: 1456–1464.
Kerscher O, Felberbaum R, Hochstrasser M . (2006). Modification of proteins by ubiquitin and ubiquitin-like proteins. Annu Rev Cell Dev Biol 22: 159–180.
Krebber H, Silver PA . (2000). Directing proteins to nucleus by fusion to nuclear localization signal tags. Methods Enzymol 327: 283–296.
Mari PO, Florea BI, Persengiev SP, Verkaik NS, Bruggenwirth HT, Modesti M et al. (2006). Dynamic assembly of end-joining complexes requires interaction between Ku70/80 and XRCC4. Proc Natl Acad Sci USA 103: 18597–18602.
Minotti G, Menna P, Salvatorelli E, Cairo G, Gianni L . (2004). Anthracyclines: molecular advances and pharmacologic developments in antitumor activity and cardiotoxicity. Pharmacol Rev 56: 185–229.
Muller S, Matunis MJ, Dejean A . (1998). Conjugation with the ubiquitin-related modifier SUMO-1 regulates the partitioning of PML within the nucleus. EMBO J 17: 61–70.
Pastink A, Eeken JC, Lohman PH . (2001). Genomic integrity and the repair of double-strand DNA breaks. Mutat Res 480–481: 37–50.
Pastwa E, Blasiak J . (2003). Non-homologous DNA end joining. Acta Biochim Pol 50: 891–908.
Prudden J, Pebernard S, Raffa G, Slavin DA, Perry JJ, Tainer JA et al. (2007). SUMO-targeted ubiquitin ligases in genome stability. EMBO J 26: 4089–4101.
Rogakou EP, Boon C, Redon C, Bonner WM . (1999). Megabase chromatin domains involved in DNA double-strand breaks in vivo. J Cell Biol 146: 905–916.
Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM . (1998). DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J Biol Chem 273: 5858–5868.
Sacher M, Pfander B, Hoege C, Jentsch S . (2006). Control of Rad52 recombination activity by double-strand break-induced SUMO modification. Nat Cell Biol 8: 1284–1290.
Sancar A, Lindsey-Boltz LA, Unsal-Kacmaz K, Linn S . (2004). Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu Rev Biochem 73: 39–85.
Seufert W, Futcher B, Jentsch S . (1995). Role of a ubiquitin-conjugating enzyme in degradation of S- and M-phase cyclins. Nature 373: 78–81.
Shayeghi M, Doe CL, Tavassoli M, Watts FZ . (1997). Characterisation of Schizosaccharomyces pombe rad31, a UBA-related gene required for DNA damage tolerance. Nucleic Acids Res 25: 1162–1169.
Song J, Durrin LK, Wilkinson TA, Krontiris TG, Chen Y . (2004). Identification of a SUMO-binding motif that recognizes SUMO-modified proteins. Proc Natl Acad Sci USA 101: 14373–14378.
Song J, Zhang Z, Hu W, Chen Y . (2005). Small ubiquitin-like modifier (SUMO) recognition of a SUMO binding motif: a reversal of the bound orientation. J Biol Chem 280: 40122–40129.
Sonoda E, Hochegger H, Saberi A, Taniguchi Y, Takeda S . (2006). Differential usage of non-homologous end-joining and homologous recombination in double strand break repair. DNA Repair (Amst) 5: 1021–1029.
Stucki M, Jackson SP . (2006). gammaH2AX and MDC1: anchoring the DNA-damage-response machinery to broken chromosomes. DNA Repair (Amst) 5: 534–543.
Sun H, Leverson JD, Hunter T . (2007). Conserved function of RNF4 family proteins in eukaryotes: targeting a ubiquitin ligase to SUMOylated proteins. EMBO J 26: 4102–4112.
Tatham MH, Jaffray E, Vaughan OA, Desterro JM, Botting C, Naismith JH et al. (2001). Polymeric chains of SUMO-2 and SUMO-3 are conjugated to protein substrates by SAE1/SAE2 and Ubc9. J Biol Chem 12: 12.
Uzunova K, Göttsche K, Miteva M, Weisshaar SR, Glanemann C, Schnellhardt M et al. (2007). Ubiquitin-dependent proteolytic control of SUMO conjugates. J Biol Chem 282: 34167–34175.
Weinstock DM, Jasin M . (2006). Alternative pathways for the repair of RAG-induced DNA breaks. Mol Cell Biol 26: 131–139.
Weinstock DM, Nakanishi K, Helgadottir HR, Jasin M . (2006). Assaying double-strand break repair pathway choice in mammalian cells using a targeted endonuclease or the RAG recombinase. Methods Enzymol 409: 524–540.
Xie Y, Kerscher O, Kroetz MB, McConchie HF, Sung P, Hochstrasser M . (2007). The yeast Hex3.Slx8 heterodimer is a ubiquitin ligase stimulated by substrate sumoylation. J Biol Chem 282: 34176–34184.
Yano K, Chen DJ . (2008). Live cell imaging of XLF and XRCC4 reveals a novel view of protein assembly in the non-homologous end-joining pathway. Cell Cycle 7: 1321–1325.
Yano K, Morotomi-Yano K, Wang SY, Uematsu N, Lee KJ, Asaithamby A et al. (2008). Ku recruits XLF to DNA double-strand breaks. EMBO Rep 9: 91–96.
Yurchenko V, Xue Z, Gama V, Matsuyama S, Sadofsky MJ . (2008). Ku70 is stabilized by increased cellular SUMO. Biochem Biophys Res Commun 366: 263–268.
Yurchenko V, Xue Z, Sadofsky MJ . (2006). SUMO modification of human XRCC4 regulates its localization and function in DNA double-strand break repair. Mol Cell Biol 26: 1786–1794.
Zhao X, Blobel G . (2005). A SUMO ligase is part of a nuclear multiprotein complex that affects DNA repair and chromosomal organization. Proc Natl Acad Sci USA 29: 4777–4782.
Acknowledgements
This work is funded by NIH Grants R01GM074748 and R01GM086171 to Y Chen, RO1CA120954 to JM Stark, R37CA050519-20 to DJ Chen and R01DE14183 to DK Ann. Y-J Li is a recipient of the NIH National Research Service Award (F32CA134180).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on the Oncogene website
Supplementary information
Rights and permissions
About this article
Cite this article
Li, YJ., Stark, J., Chen, D. et al. Role of SUMO:SIM-mediated protein–protein interaction in non-homologous end joining. Oncogene 29, 3509–3518 (2010). https://doi.org/10.1038/onc.2010.108
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/onc.2010.108
Keywords
This article is cited by
-
Post-translational modifications: emerging regulators manipulating jasmonate biosynthesis and signaling
Plant Cell Reports (2022)