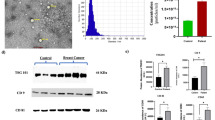
Abstract
Ductal carcinoma in situ (DCIS) of the breast is part of a spectrum of preinvasive lesions that originate within normal breast tissue and progress to invasive breast cancer. The detection of DCIS is important for the reduction of mortality from breast cancer, but the diagnosis of preinvasive breast tumors is hampered by the lack of an adequate detection method. To identify the changes in protein expression during the initial stage of tumorigenesis and to identify the presence of new DCIS markers, we analysed serum from 60 patients with breast cancer and 60 normal controls using mass spectrometry. A 23-protein index was generated that correctly distinguishes the DCIS and control groups with sensitivities and specificities in excess of 80% in two independent cohorts. Two candidate peptides were purified and identified as platelet factor 4 (PF-4) and complement C3adesArg anaphylatoxin (C3adesArg) using liquid chromatography–tandem mass spectrometry (LC–MS/MS). In an independent serum set of 165 patients, PF-4 and C3adesArg were significantly upregulated in DCIS compared with non-cancerous controls, as validated using western blot and enzyme-linked immunosorbent assay. We conclude that our serum protein-based test, used in conjunction with image-based screening practices, could improve the sensitivity and specificity of breast cancer detection.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Albrethsen J . (2007). Reproducibility in protein profiling by MALDI-TOF mass spectrometry. Clin Chem 53: 852–858.
Alexander H, Stegner AL, Wagner-Mann C, Du Bois GC, Alexander S, Sauter ER . (2004). Proteomic analysis to identify breast cancer biomarkers in nipple aspirate fluid. Clin Cancer Res 10: 7500–7510.
Becker S, Cazares LH, Watson P, Lynch H, Semmes OJ, Drake RR et al. (2004). Surfaced-enhanced laser desorption/ionization time-of-flight (SELDI-TOF) differentiation of serum protein profiles of BRCA-1 and sporadic breast cancer. Ann Surg Oncol 11: 907–914.
Belluco C, Petricoin EF, Mammano E, Facchiano F, Ross-Rucker S, Nitti D et al. (2007). Serum proteomic analysis identifies a highly sensitive and specific discriminatory pattern in stage 1 breast cancer. Ann Surg Oncol 14: 2470–2476.
Bijker N, Meijnen P, Peterse JL, Bogaerts J, Van Hoorebeeck I, Julien JP et al. (2006). Breast-conserving treatment with or without radiotherapy in ductal carcinoma-in-situ: ten-year results of European Organisation for Research and Treatment of Cancer randomized phase III trial 10853--a study by the EORTC Breast Cancer Cooperative Group and EORTC Radiotherapy Group. J Clin Oncol 24: 3381–3387.
Bjorge L, Hakulinen J, Vintermyr OK, Jarva H, Jensen TS, Iversen OE et al. (2005). Ascitic complement system in ovarian cancer. Br J Cancer 92: 895–905.
Brozkova K, Budinska E, Bouchal P, Hernychova L, Knoflickova D, Valik D et al. (2008). Surface-enhanced laser desorption/ionization time-of-flight proteomic profiling of breast carcinomas identifies clinicopathologically relevant groups of patients similar to previously defined clusters from cDNA expression. Breast Cancer Res 10: R48.
Carlevaro MF, Albini A, Ribatti D, Gentili C, Benelli R, Cermelli S et al. (1997). Transferrin promotes endothelial cell migration and invasion: implication in cartilage neovascularization. J Cell Biol 136: 1375–1384.
Cavanaugh PG, Jia L, Zou Y, Nicolson GL . (1999). Transferrin receptor overexpression enhances transferrin responsiveness and the metastatic growth of a rat mammary adenocarcinoma cell line. Breast Cancer Res Treat 56: 203–217.
Cavanaugh PG, Nicolson GL . (1998). Selection of highly metastatic rat MTLn2 mammary adenocarcinoma cell variants using in vitro growth response to transferrin. J Cell Physiol 174: 48–57.
Cervi D, Yip TT, Bhattacharya N, Podust VN, Peterson J, Abou-Slaybi A et al. (2008). Platelet-associated PF-4 as a biomarker of early tumor growth. Blood 111: 1201–1207.
Cho WC . (2007). Contribution of oncoproteomics to cancer biomarker discovery. Mol Cancer 6: 25.
Elmore JG, Armstrong K, Lehman CD, Fletcher SW . (2005). Screening for breast cancer. JAMA 293: 1245–1256.
Esserman LJ, Shieh Y, Park JW, Ozanne EM . (2007). A role for biomarkers in the screening and diagnosis of breast cancer in younger women. Expert Rev Mol Diagn 7: 533–544.
Gast MC, Bonfrer JM, van Dulken EJ, de Kock L, Rutgers EJ, Schellens JH et al. (2006). SELDI-TOF MS serum protein profiles in breast cancer: assessment of robustness and validity. Cancer Biomark 2: 235–248.
Gast MC, van Tinteren H, Bontenbal M, van Hoesel RQ, Nooij MA, Rodenhuis S et al. (2008). Haptoglobin phenotype is not a predictor of recurrence free survival in high-risk primary breast cancer patients. BMC Cancer 8: 389.
Gengrinovitch S, Greenberg SM, Cohen T, Gitay-Goren H, Rockwell P, Maione TE et al. (1995). Platelet factor-4 inhibits the mitogenic activity of VEGF121 and VEGF165 using several concurrent mechanisms. J Biol Chem 270: 15059–15065.
Goncalves A, Esterni B, Bertucci F, Sauvan R, Chabannon C, Cubizolles M et al. (2006). Postoperative serum proteomic profiles may predict metastatic relapse in high-risk primary breast cancer patients receiving adjuvant chemotherapy. Oncogene 25: 981–989.
Greene FL, Page DL, Fleming ID et al. (2002). Cancer Staging Manual, 6th edn. Springer: New York.
Gupta SK, Hassel T, Singh JP . (1995). A potent inhibitor of endothelial cell proliferation is generated by proteolytic cleavage of the chemokine platelet factor 4. Proc Natl Acad Sci USA 92: 7799–7803.
Gupta SK, Singh JP . (1994). Inhibition of endothelial cell proliferation by platelet factor-4 involves a unique action on S phase progression. J Cell Biol 127: 1121–1127.
Hagedorn M, Zilberberg L, Lozano RM, Cuevas P, Canron X, Redondo-Horcajo M et al. (2001). A short peptide domain of platelet factor 4 blocks angiogenic key events induced by FGF-2. FASEB J 15: 550–552.
Hanash S . (2003). Disease proteomics. Nature 422: 226–232.
Hanash SM, Pitteri SJ, Faca VM . (2008). Mining the plasma proteome for cancer biomarkers. Nature 452: 571–579.
Harris L, Fritsche H, Mennel R, Norton L, Ravdin P, Taube S et al. (2007). American Society of Clinical Oncology 2007 update of recommendations for the use of tumor markers in breast cancer. J Clin Oncol 25: 5287–5312.
Hu Y, Zhang S, Yu J, Liu J, Zheng S . (2005). SELDI-TOF-MS: the proteomics and bioinformatics approaches in the diagnosis of breast cancer. Breast 14: 250–255.
Jacot W, Lhermitte L, Dossat N, Pujol JL, Molinari N, Daures JP et al. (2008). Serum proteomic profiling of lung cancer in high-risk groups and determination of clinical outcomes. J Thorac Oncol 3: 840–850.
Jemal A, Siegel R, Ward E, Hao Y, Xu J, Murray T et al. (2008). Cancer statistics, 2008. CA Cancer J Clin 58: 71–96.
Kang X, Xu Y, Wu X, Liang Y, Wang C, Guo J et al. (2005). Proteomic fingerprints for potential application to early diagnosis of severe acute respiratory syndrome. Clin Chem 51: 56–64.
Kopczynski Z, Kuzniak J, Thielemann A, Kaczmarek J, Rybczynska M . (1998). The biochemical modification of the erythrocyte membranes from women with ovarian cancer. Br J Cancer 78: 466–471.
Kozak KR, Su F, Whitelegge JP, Faull K, Reddy S, Farias-Eisner R . (2005). Characterization of serum biomarkers for detection of early stage ovarian cancer. Proteomics 5: 4589–4596.
Kramar A, Faraggi D, Fortune A, Reiser B . (2001). mROC: a computer program for combining tumour markers in predicting disease states. Comput Methods Programs Biomed 66: 199–207.
Kuerer HM, Albarracin CT, Yang WT, Cardiff RD, Brewster AM, Symmans WF et al. (2009). Ductal carcinoma in situ: state of the science and roadmap to advance the field. J Clin Oncol 27: 279–288.
Lee IN, Chen CH, Sheu JC, Lee HS, Huang GT, Chen DS et al. (2006). Identification of complement C3a as a candidate biomarker in human chronic hepatitis C and HCV-related hepatocellular carcinoma using a proteomics approach. Proteomics 6: 2865–2873.
Levenson VV . (2007). Biomarkers for early detection of breast cancer: what, when, and where? Biochim Biophys Acta 1770: 847–856.
Li J, Orlandi R, White CN, Rosenzweig J, Zhao J, Seregni E et al. (2005). Independent validation of candidate breast cancer serum biomarkers identified by mass spectrometry. Clin Chem 51: 2229–2235.
Li J, Zhang Z, Rosenzweig J, Wang YY, Chan DW . (2002). Proteomics and bioinformatics approaches for identification of serum biomarkers to detect breast cancer. Clin Chem 48: 1296–1304.
Maione TE, Gray GS, Petro J, Hunt AJ, Donner AL, Bauer SI et al. (1990). Inhibition of angiogenesis by recombinant human platelet factor-4 and related peptides. Science 247: 77–79.
Mange A, Bellet V, Tuaillon E, Van de Perre P, Solassol J . (2008). Comprehensive proteomic analysis of the human milk proteome: contribution of protein fractionation. J Chromatogr B Analyt Technol Biomed Life Sci 876: 252–256.
Mathelin C, Cromer A, Wendling C, Tomasetto C, Rio MC . (2006). Serum biomarkers for detection of breast cancers: A prospective study. Breast Cancer Res Treat 96: 83–90.
Miguet L, Bogumil R, Decloquement P, Herbrecht R, Potier N, Mauvieux L et al. (2006). Discovery and identification of potential biomarkers in a prospective study of chronic lymphoid malignancies using SELDI-TOF-MS. J Proteome Res 5: 2258–2269.
Nicolson GL, Cavanaugh PG, Inoue T . (1992). Differential stimulation of the growth of lung-metastasizing tumor cells by lung (paracrine) growth factors: identification of transferrin-like mitogens in lung tissue-conditioned medium. J Natl Cancer Inst Monogr 13: 153–161.
Orel SG, Kay N, Reynolds C, Sullivan DC . (1999). BI-RADS categorization as a predictor of malignancy. Radiology 211: 845–850.
Pavlakis K, Messini I, Vrekoussis T, Yiannou P, Keramopoullos D, Louvrou N et al. (2008). The assessment of angiogenesis and fibroblastic stromagenesis in hyperplastic and pre-invasive breast lesions. BMC Cancer 8: 88.
Perollet C, Han ZC, Savona C, Caen JP, Bikfalvi A . (1998). Platelet factor 4 modulates fibroblast growth factor 2 (FGF-2) activity and inhibits FGF-2 dimerization. Blood 91: 3289–3299.
Petricoin EF, Liotta LA . (2004). SELDI-TOF-based serum proteomic pattern diagnostics for early detection of cancer. Curr Opin Biotechnol 15: 24–30.
Pierce BL, Ballard-Barbash R, Bernstein L, Baumgartner RN, Neuhouser ML, Wener MH et al. (2009). Elevated biomarkers of inflammation are associated with reduced survival among breast cancer patients. J Clin Oncol 27: 3437–3444.
Rai AJ, Zhang Z, Rosenzweig J, Shih Ie M, Pham T, Fung ET et al. (2002). Proteomic approaches to tumor marker discovery. Arch Pathol Lab Med 126: 1518–1526.
Roesch-Ely M, Nees M, Karsai S, Ruess A, Bogumil R, Warnken U et al. (2007). Proteomic analysis reveals successive aberrations in protein expression from healthy mucosa to invasive head and neck cancer. Oncogene 26: 54–64.
Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N et al. (2003). TM4: a free, open-source system for microarray data management and analysis. Biotechniques 34: 374–378.
Sahu A, Lambris JD . (2001). Structure and biology of complement protein C3, a connecting link between innate and acquired immunity. Immunol Rev 180: 35–48.
Schagger H . (2006). Tricine-SDS-PAGE. Nat Protoc 1: 16–22.
Semmes OJ, Feng Z, Adam BL, Banez LL, Bigbee WL, Campos D et al. (2005). Evaluation of serum protein profiling by surface-enhanced laser desorption/ionization time-of-flight mass spectrometry for the detection of prostate cancer: I. Assessment of platform reproducibility. Clin Chem 51: 102–112.
Sharpe RJ, Byers HR, Scott CF, Bauer SI, Maione TE . (1990). Growth inhibition of murine melanoma and human colon carcinoma by recombinant human platelet factor 4. J Natl Cancer Inst 82: 848–853.
Solassol J, Jacot W, Lhermitte L, Boulle N, Maudelonde T, Mange A . (2006). Clinical proteomics and mass spectrometry profiling for cancer detection. Expert Rev Proteomics 3: 311–320.
Solassol J, Marin P, Demettre E, Rouanet P, Bockaert J, Maudelonde T et al. (2005). Proteomic detection of prostate-specific antigen using a serum fractionation procedure: potential implication for new low-abundance cancer biomarkers detection. Anal Biochem 338: 26–31.
Song J, Patel M, Rosenzweig CN, Chan-Li Y, Sokoll LJ, Fung ET et al. (2006). Quantification of fragments of human serum inter-alpha-trypsin inhibitor heavy chain 4 by a surface-enhanced laser desorption/ionization-based immunoassay. Clin Chem 52: 1045–1053.
Timms JF, Arslan-Low E, Gentry-Maharaj A, Luo Z, T'Jampens D, Podust VN et al. (2007). Preanalytic influence of sample handling on SELDI-TOF serum protein profiles. Clin Chem 53: 645–656.
Van der Merwe DE, Oikonomopoulou K, Marshall J, Diamandis EP . (2007). Mass spectrometry: uncovering the cancer proteome for diagnostics. Adv Cancer Res 96: 23–50.
Vlahou A, Laronga C, Wilson L, Gregory B, Fournier K, McGaughey D et al. (2003). A novel approach toward development of a rapid blood test for breast cancer. Clin Breast Cancer 4: 203–209.
Ward DG, Suggett N, Cheng Y, Wei W, Johnson H, Billingham LJ et al. (2006). Identification of serum biomarkers for colon cancer by proteomic analysis. Br J Cancer 94: 1898–1905.
Acknowledgements
This work was supported in part by grants from the Comité de l'Hérault de la Ligue Contre le Cancer. Serum collection was supported by INSERM (RBM 03-63).
Author information
Authors and Affiliations
Corresponding author
Additional information
Supplementary Information accompanies the paper on the Oncogene website (http://www.nature.com/onc)
Supplementary information
Rights and permissions
About this article
Cite this article
Solassol, J., Rouanet, P., Lamy, P. et al. Serum protein signature may improve detection of ductal carcinoma in situ of the breast. Oncogene 29, 550–560 (2010). https://doi.org/10.1038/onc.2009.341
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/onc.2009.341
Keywords
This article is cited by
-
Novel serum protein biomarker panel revealed by mass spectrometry and its prognostic value in breast cancer
Breast Cancer Research (2014)
-
Identification of Potential Markers Related to Neoadjuvant Chemotherapy Sensitivity of Breast Cancer by SELDI-TOF MS
Applied Biochemistry and Biotechnology (2012)
-
Early diagnostic protein biomarkers for breast cancer: how far have we come?
Breast Cancer Research and Treatment (2012)