Abstract
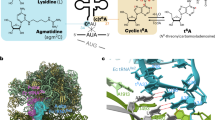
In the 2.7-Å resolution crystal structure of methionyl-tRNA synthetase (MetRS) in complex with tRNAMet and a methionyl-adenylate analog, the tRNA anticodon loop is distorted to form a triple-base stack comprising C34, A35 and A38. A tryptophan residue stacks on C34 to extend the triple-base stack. In addition, C34 forms Watson-Crick–type hydrogen bonds with Arg357. This structure resolves the longstanding question of how MetRS specifically recognizes tRNAMet.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Arnez, J.G. & Moras, D. Trends Biochem. Sci. 22, 211–216 (1997).
Schulman, L.H. & Pelka, H. Science 242, 765–768 (1988).
Muramatsu, T. et al. Nature 336, 179–181 (1988).
Ghosh, G., Pelka, H. & Schulman, L.H. Biochemistry 29, 2220–2225 (1990).
Hauenstein, S., Zhang, C.M., Hou, Y.M. & Perona, J.J. Nat. Struct. Mol. Biol. 11, 1134–1141 (2004).
Fukai, S. et al. RNA 9, 100–111 (2003).
Silvian, L.F., Wang, J. & Steitz, T.A. Science 285, 1074–1077 (1999).
Cusack, S., Yaremchuk, A. & Tukalo, M. EMBO J. 19, 2351–2361 (2000).
Cavarelli, J., Delagoutte, B., Eriani, G., Gangloff, J. & Moras, D. EMBO J. 17, 5438–5448 (1998).
Mechulam, Y. et al. J. Mol. Biol. 294, 1287–1297 (1999).
Sugiura, I. et al. Structure 8, 197–208 (2000).
Crepin, T., Schmitt, E., Blanquet, S. & Mechulam, Y. Biochemistry 43, 2635–2644 (2004).
Ghosh, G., Kim, H.Y., Demaret, J.P., Brunie, S. & Schulman, L.H. Biochemistry 30, 11767–11774 (1991).
Mellot, P., Mechulam, Y., Le Corre, D., Blanquet, S. & Fayat, G. J. Mol. Biol. 208, 429–443 (1989).
Nakanishi, K. et al. Proc. Natl. Acad. Sci. USA 102, 7487–7492 (2005).
Acknowledgements
We thank M. Kawamoto and N. Shimizu (Japan Synchrotron Radiation Research Institute) for their help in data collection at SPring-8. This work was supported by a grant from the Ministry of Education, Culture, Sports, Science and Technology to O.N., by a Precursory Research for Embryonic Science and Technology Program grant from Japan Science and Technology and by Sumitomo and Naito Foundation grants to O.N.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Fig. 1
MetSA bound to the amino-acylation site of MetRS (PDF 643 kb)
Supplementary Fig. 2
Conformational change of MetRS upon tRNAMetm binding (PDF 849 kb)
Supplementary Fig. 3
The 2|Fo|-|Fc| simulated annealing omit map (PDF 1173 kb)
Supplementary Fig. 4
Sequence alignment of MetRSs (PDF 1053 kb)
Supplementary Fig. 5
Comparison of tRNA anticodon recognition by the class Ia aaRSs (PDF 1853 kb)
Supplementary Table 1
Data collection and refinement statistics (PDF 63 kb)
Rights and permissions
About this article
Cite this article
Nakanishi, K., Ogiso, Y., Nakama, T. et al. Structural basis for anticodon recognition by methionyl-tRNA synthetase. Nat Struct Mol Biol 12, 931–932 (2005). https://doi.org/10.1038/nsmb988
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb988