Abstract
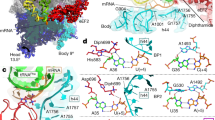
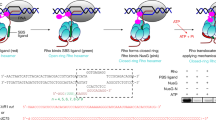
In bacteria, incorporation of selenocysteine, the 21st amino acid, into proteins requires elongation factor SelB, which has the unusual property of binding to both transfer RNA (tRNA) and mRNA. SelB binds to an mRNA hairpin formed by the selenocysteine insertion sequence (SECIS) with extremely high specificity, the molecular basis of which has been unknown. We have determined the crystal structure of the mRNA-binding domain of SelB in complex with SECIS RNA at a resolution of 2.3 Å. This is the first example of a complex between an RNA and a winged-helix (WH) domain, a motif found in many DNA-binding proteins and recently discovered in RNA-binding proteins. Notably, RNA binding does not induce a major conformational change in the WH motif. The structure reveals a new mode of RNA recognition with a geometry that allows the complex to wrap around the small ribosomal subunit.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Böck, A. et al. Selenocysteine: the 21st amino acid. Mol. Microbiol. 5, 515–520 (1991).
Boyington, J.C., Gladyshev, V.N., Khangulov, S.V., Stadtman, T.C. & Sun, P.D. Crystal structure of formate dehydrogenase H: catalysis involving Mo, molybdopterin, selenocysteine, and an Fe4S4 cluster. Science 275, 1305–1308 (1997).
Forchhammer, K., Leinfelder, W. & Böck, A. Identification of a novel translation factor necessary for the incorporation of selenocysteine into protein. Nature 342, 453–456 (1989).
Nissen, P. et al. Crystal structure of the ternary complex of Phe-tRNAPhe, EF-Tu and a GTP analog. Science 270, 1464–1472 (1995).
Stark, H. et al. Visualization of elongation factor Tu on the Escherichia coli ribosome. Nature 389, 403–406 (1997).
Pape, T., Wintermeyer, W. & Rodnina, M. Induced fit in initial selection and proofreading of aminoacyl-tRNA on the ribosome. EMBO J. 18, 3800–3807 (1999).
Ogle, J.M., Murphy, F.V., Tarry, M.J. & Ramakrishnan, V. Selection of tRNA by the ribosome requires a transition from an open to a closed form. Cell 111, 721–732 (2002).
Kromayer, M., Wilting, R., Tormay, P. & Böck, A. Domain structure of the prokaryotic selenocysteine-specific elongation factor SelB. J. Mol. Biol. 262, 413–420 (1996).
Thanbichler, M., Böck, A. & Goody, R.S. Kinetics of the interaction of translation factor SelB from Escherichia coli with guanosine nucleotides and selenocysteine insertion sequence RNA. J. Biol. Chem. 275, 20458–20466 (2000).
Heider, J., Baron, C. & Böck, A. Coding from a distance: dissection of the mRNA determinants required for the incorporation of selenocysteine into protein. EMBO J. 11, 3759–3766 (1992).
Li, C., Reches, M. & Engelberg-Kulka, H. The bulged nucleotide in the Escherichia coli minimal selenocysteine insertion sequence participates in interaction with SelB: a genetic approach. J. Bacteriol. 182, 6302–6307 (2000).
Fourmy, D., Guittet, E. & Yoshizawa, S. Structure of prokaryotic SECIS mRNA hairpin and its interaction with elongation factor SelB. J. Mol. Biol. 324, 137–150 (2002).
Hüttenhofer, A., Westhof, E. & Böck, A. Solution structure of mRNA hairpins promoting selenocysteine incorporation in Escherichia coli and their base-specific interaction with special elongation factor SELB. RNA 2, 354–366 (1996).
Sandman, K.E., Tardiff, D.F., Neely, L.A. & Noren, C.J. Revised Escherichia coli selenocysteine insertion requirements determined by in vivo screening of combinatorial libraries of SECIS variants. Nucleic Acids Res. 31, 2234–2241 (2003).
Selmer, M. & Su, X.D. Crystal structure of an mRNA-binding fragment of Moorella thermoacetica elongation factor SelB. EMBO J. 21, 4145–4153 (2002).
Wintjens, R. & Rooman, M. Structural classification of HTH DNA-binding domains and protein-DNA interaction modes. J. Mol. Biol. 262, 294–313 (1996).
Clark, K.L., Halay, E.D., Lai, E. & Burley, S.K. Co-crystal structure of the HNF-3/fork head DNA-recognition motif resembles histone H5. Nature 364, 412–420 (1993).
Gajiwala, K.S. & Burley, S.K. Winged helix proteins. Curr. Opin. Struct. Biol. 10, 110–116 (2000).
Hüttenhofer, A. & Böck, A. Selenocysteine inserting RNA elements modulate GTP hydrolysis of elongation factor SelB. Biochemistry 37, 885–890 (1998).
Batey, R.T., Rambo, R.P., Lucast, L., Rha, B. & Doudna, J.A. Crystal structure of the ribonucleoprotein core of the signal recognition particle. Science 287, 1232–1239 (2000).
Alfano, C. et al. Structural analysis of cooperative RNA binding by the La motif and central RRM domain of human La protein. Nat. Struct. Mol. Biol. 11, 323–329 (2004).
Dong, G., Chakshusmathi, G., Wolin, S.L. & Reinisch, K.M. Structure of the La motif: a winged helix domain mediates RNA binding via a conserved aromatic patch. EMBO J. 23, 1000–1007 (2004).
Schumacher, M., Lau, A. & Johnson, P. Structural basis of core promoter recognition in a primitive eukaryote. Cell 115, 413–424 (2003).
Gajiwala, K.S. et al. Structure of the winged-helix protein hRFX1 reveals a new mode of DNA binding. Nature 403, 916–921 (2000).
Schwartz, T., Rould, M.A., Lowenhaupt, K., Herbert, A. & Rich, A. Crystal structure of the Zα domain of the human editing enzyme ADAR1 bound to left-handed Z-DNA. Science 284, 1841–1845 (1999).
Ryder, S.P., Ortoleva-Donelly, L., Kosek, A.B. & Strobel, S.A. Chemical probing of RNA by nucleotide analog interference mapping. Methods Enzymol. 317, 92–109 (2000).
Mo, Y., Vaessen, B., Johnston, K. & Marmorstein, R. Structures of SAP-1 bound to DNA targets from the E74 and c-fos promoters: insights into DNA sequence discrimination by Ets proteins. Mol. Cell 2, 201–212 (1998).
Yusupova, G.Z., Yusupov, M.M., Cate, J.H. & Noller, H.F. The path of messenger RNA through the ribosome. Cell 106, 233–241 (2001).
Valegard, K., Murray, J.B., Stockley, P.G., Stonehouse, N.J. & Liljas, L. Crystal structure of an RNA bacteriophage coat protein–operator complex. Nature 371, 623–626 (1994).
Oubridge, C., Ito, N., Evans, P.R., Teo, C.H. & Nagai, K. Crystal structure at 1.92 Å resolution of the RNA-binding domain of the U1A spliceosomal protein complexed with an RNA hairpin. Nature 372, 432–438 (1994).
Allain, F.H. et al. Specificity of ribonucleoprotein interaction determined by RNA folding during complex formulation. Nature 380, 646–650 (1996).
Rould, M.A., Perona, J.J. & Steitz, T.A. Structural basis of anticodon loop recognition by glutaminyl-tRNA synthetase. Nature 352, 213–218 (1991).
Cavarelli, J., Rees, B., Ruff, M., Thierry, J.C. & Moras, D. Yeast tRNA(Asp) recognition by its cognate class II aminoacyl-tRNA synthetase. Nature 362, 181–184 (1993).
Cusack, S., Yaremchuk, A., Krikliviy, I. & Tukalo, M. tRNA(Pro) anticodon recognition by Thermus thermophilus prolyl-tRNA synthetase. Structure 6, 101–108 (1998).
Kromayer, M., Neuhierl, B., Friebel, A. & Bock, A. Genetic probing of the interaction between the translation factor SelB and its mRNA binding element in Escherichia coli. Mol. Gen. Genet. 262, 800–806 (1999).
Klug, S.J., Huttenhofer, A., Kromayer, M. & Famulok, M. In vitro and in vivo characterization of novel mRNA motifs that bind special elongation factor SelB. Proc. Natl. Acad. Sci. USA 94, 6676–6681 (1997).
Smith, D.B. & Johnson, K.S. Single-step purification of polypeptides expressed in Escherichia coli as fusions with glutathione S-transferase. Gene 67, 31–40 (1988).
Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997).
Collaborative Computational Project, Number 4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D 50, 760–763 (1994).
Brunger, A.T. et al. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D 54, 905–921 (1998).
Jones, T.A., Zou, J.Y., Cowan, S.W. & Kjeldgaard, M. Improved methods for building protein models in electron density maps and the locations of errors in these methods. Acta Crystallogr. A 47, 110–119 (1991).
Laskowski, R., MacArthur, M.W., Moss, D.S. & Thornton, J.M. PROCHECK: a program to check the streochemical quality of protein structures. J. Appl. Cryst. 26, 283–291 (1993).
Blanchard, S.C., Fourmy, D., Eason, R.G. & Puglisi, J.D. rRNA chemical groups required for aminoglycoside binding. Biochemistry 37, 7716–7724 (1998).
Stern, S., Moazed, D. & Noller, H.F. Structural analysis of RNA using chemical and enzymatic probing monitored by primer extension. Methods Enzymol. 164, 481–489 (1988).
Schwartz, T., Behlke, J., Lowenhaupt, K., Heinemann, U. & Rich, A. Structure of the DLM-1–Z-DNA complex reveals a conserved family of Z-DNA-binding proteins. Nat. Struct. Biol. 8, 761–765 (2001).
Wimberly, B.T. et al. Structure of the 30S ribosomal subunit. Nature 407, 327–339 (2000).
Acknowledgements
We thank M. Kawamoto, H. Sakai and K. Miura for assistance in data collection at SPring-8. This work was supported by the joint program JSPS-CNRS to S.Y. and K.M. and the French national program GEOMEX to S.Y. and D.F. K.M. and D.K. were supported in part by the Ministry of Education, Science, Sports, Culture and Technology of Japan, and the Protein 3000 project. L.R. was supported by a Japan Society for the Promotion of Science postdoctoral fellowship for foreign researchers.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Fig. 1
Crystal packing of SelB-M–RNA complexes. (PDF 249 kb)
Supplementary Fig. 2
Structure-based sequence alignment of bacterial SelB mRNA-binding domains. (PDF 464 kb)
Supplementary Fig. 3
Protein-RNA interactions in the SelB-M–SECIS RNA complex. (PDF 343 kb)
Supplementary Fig. 4
Overview of the protein-RNA loop interactions. (PDF 1859 kb)
Rights and permissions
About this article
Cite this article
Yoshizawa, S., Rasubala, L., Ose, T. et al. Structural basis for mRNA recognition by elongation factor SelB. Nat Struct Mol Biol 12, 198–203 (2005). https://doi.org/10.1038/nsmb890
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb890
This article is cited by
-
De novo computational RNA modeling into cryo-EM maps of large ribonucleoprotein complexes
Nature Methods (2018)
-
Genetic code flexibility in microorganisms: novel mechanisms and impact on physiology
Nature Reviews Microbiology (2015)
-
Roquin binding to target mRNAs involves a winged helix-turn-helix motif
Nature Communications (2014)
-
Effects of combinatorial expression of selA, selB and selC genes on the efficiency of selenocysteine incorporation in Escherichia coli
Chemical Research in Chinese Universities (2013)
-
Nprl3 is required for normal development of the cardiovascular system
Mammalian Genome (2012)