Abstract
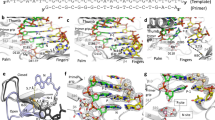
Tenofovir, also known as PMPA, R-9-(2-(phosphonomethoxypropyl)adenine, is a nucleotide reverse transcriptase (RT) inhibitor. We have determined the crystal structures of two related complexes of HIV-1 RT with template primer and tenofovir: (i) a ternary complex at a resolution of 3.0 Å of RT crosslinked to a dideoxy-terminated DNA with tenofovir-diphosphate bound as the incoming substrate; and (ii) a RT–DNA complex at a resolution of 3.1 Å with tenofovir at the 3′ primer terminus. The tenofovir nucleotide in the tenofovir-terminated structure seems to adopt multiple conformations. Some nucleoside reverse transcriptase inhibitors, including 3TC and AZT, have elements ('handles') that project beyond the corresponding elements on normal dNTPs (the 'substrate envelope'). HIV-1 RT resistance mechanisms to AZT and 3TC take advantage of these handles; tenofovir's structure lacks handles that could protrude through the substrate envelope to cause resistance.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Huang, H., Chopra, R., Verdine, G.L. & Harrison, S.C. Structure of a covalently trapped catalytic complex of HIV-1 reverse transcriptase: implications for drug resistance. Science 282, 1669– 1675 (1998).
Sarafianos, S.G. et al. Lamivudine (3TC) resistance in HIV-1 reverse transcriptase involves steric hindrance with β-branched amino acids. Proc. Natl. Acad. Sci. USA 96, 10027– 10032 (1999).
Gao, H.Q., Boyer, P.L., Sarafianos, S.G., Arnold, E. & Hughes, S.H. The role of steric hindrance in 3TC resistance of human immunodeficiency virus type-1 reverse transcriptase. J. Mol. Biol. 300, 403– 418 (2000).
Boyer, P.L. et al. YADD mutants of human immunodeficiency virus type 1 and Moloney murine leukemia virus reverse transcriptase are resistant to lamivudine triphosphate (3TCTP) in vitro . J. Virol. 75, 6321– 6328 (2001).
Arion, D., Kaushik, N., McCormick, S., Borkow, G. & Parniak, M.A. Phenotypic mechanism of HIV-1 resistance to 3_-azido-3_-deoxythymidine (AZT): increased polymerization processivity and enhanced sensitivity to pyrophosphate of the mutant viral reverse transcriptase. Biochemistry 37, 15908– 15917 (1998).
Meyer, P.R., Matsuura, S.E., Mian, A.M., So, A.G. & Scott, W.A. A mechanism of AZT resistance: an increase in nucleotide-dependent primer unblocking by mutant HIV-1 reverse transcriptase. Mol. Cell 4, 35– 43 (1999).
Boyer, P.L., Sarafianos, S.G., Arnold, E. & Hughes, S.H. Selective excision of AZTMP by drug-resistant human immunodeficiency virus reverse transcriptase. J. Virol. 75, 4832– 4842 (2001).
Boyer, P.L., Sarafianos, S.G., Arnold, E. & Hughes, S.H. The M184V mutation reduces the selective excision of zidovudine 5_-monophosphate (AZTMP) by the reverse transcriptase of human immunodeficiency virus type 1. J. Virol. 76, 3248– 3256 (2002).
Srinivas, R. & Fridland, A. Antiviral activities of 9-R-2 phosphonomethoxypropyl adenine (PMPA) and bis(isopropyloxymethylcarbonyl)PMPA against various drug-resistant human immunodeficiency virus strains. Antimicrob. Agents Chemother. 42, 1484– 1487 (1998).
Wainberg, M.A. et al. In vitro selection and characterization of HIV-1 with reduced susceptibility to PMPA. Antivir. Ther. 4, 87– 94 (1999).
Margot N.A., et al. Genotypic and phenotypic analyses of HIV-1 in antiretroviral-experienced patients treated with tenofovir DF. AIDS 16, 1227– 1235 (2002).
Jacobo-Molina, A. et al. Crystal structure of human immunodeficiency virus type 1 reverse transcriptase complexed with double-stranded DNA at 3.0 Å resolution shows bent DNA. Proc. Natl. Acad. Sci. USA 90, 6320– 6324 (1993).
Ding, J. et al. Structure and functional implications of the polymerase active site region in a complex of HIV-1 RT with double-stranded DNA and an antibody Fab fragment at 2.8 Å resolution. J. Mol. Biol. 284, 1095– 1111 (1998).
Sarafianos, S.G. et al. Crystal structure of HIV-1 reverse transcriptase in complex with a polypurine tract RNA:DNA. EMBO J. 20, 1449– 1461 (2001).
Sarafianos, S.G. et al. Structures of HIV-1 reverse transcriptase with pre-and post-translocation AZTMP-terminated DNA. EMBO J. 21, 6614– 6624 (2002).
Doublie, S., Tabor, S., Long, A.M., Richardson, C.C. & Ellenberger, T. Crystal structure of a bacteriophage T7 DNA replication complex at 2.2 Å resolution. Nature 391, 251– 258 (1998).
Li, Y., Korolev, S. & Waksman, G. Crystal structures of open and closed forms of binary and ternary complexes of the large fragment of Thermus aquaticus DNA polymerase: structural basis for nucleotide incorporation. EMBO J. 17, 7514– 7525 (1998).
Dahlberg, M.E. & Benkovic, S.J. Kinetic mechanism of DNA polymerase I (Klenow fragment): identification of a second conformational change. Biochemistry 30, 4835– 4843 (1991).
Hsieh, J., Zinnen, S. & Modrich, P. Kinetic mechanism of the DNA-dependent DNA polymerase activity of human immunodeficiency virus reverse transcriptase. J. Biol. Chem. 268, 24607– 24613 (1993).
Kati, W.M., Johnson, K.A., Jerva, L.F. & Anderson, K.S. Mechanism and fidelity of HIV reverse transcriptase. J. Biol. Chem. 267, 25988– 25997 (1992).
Suo, Z. & Johnson, K.A. Selective inhibition of HIV-1 reverse transcriptase by an antiviral inhibitor (R)-9-(2-phosphonylmethoxypropyl)adenine. J. Biol. Chem. 273, 27250– 27258 (1998).
Prabu-Jeyabalan, M., Nalivaika, E. & Schiffer, C.A. Substrate shape determines specificity of recognition for HIV-1 protease: analysis of crystal structures of six substrate complexes. Structure 10, 369– 381 (2002).
White, K.L. et al. Molecular mechanisms of resistance to human immunodeficiency virus type 1 with reverse transcriptase mutations K65R and K65R+M184V and their effects on enzyme function and viral replication capacity. Antimicrob. Agents Chemother. 46, 3437– 3446 (2002).
Selmi, B., Boretto, J., Sarfati, S.R., Guerreiro, C. & Canard, B. Mechanism-based suppression of dideoxynucleotide resistance by K65R human immunodeficiency virus reverse transcriptase using an α-boranophosphate nucleoside analogue. J. Biol. Chem. 276, 48466– 48472 (2001).
Arion, D., Borkow, G., Gu, Z., Wainberg, M.A. & Parniak, M.A. The K65R mutation confers increased DNA polymerase processivity to HIV-1 reverse transcriptase. J. Biol. Chem. 271, 19860– 19864 (1996).
Miller, M.D. et al. Characterization of resistance mutation patterns emerging over two years during first line antiretroviral treatment with tenofovir DF or stavudine in combination with lamivudine and efavirenz. Antivir. Ther. 8, S1– S185 (2003).
Boyer, P.L., Sarafianos, S., Arnold, E. & Hughes, S.H. Nucleoside analog resistance caused by insertions in the fingers of human immunodeficiency virus type1 reverse transcriptase involves ATP-mediated excision. J. Virol. 76, 9143– 9151 (2002).
Harrigan, P.R., Miller, M.D., McKenna, P., Brumme, Z.L. & Larder, B.A. Phenotypic susceptibilities to tenofovir in a large panel of clinically derived human immunodeficiency virus type 1 isolates. Antimicrob. Agents Chemother. 46, 1067– 1072 (2002).
Mas, A. et al. Role of a dipeptide insertion between codons 69 and 70 of HIV-1 reverse transcriptase in the mechanism of AZT resistance. EMBO J. 19, 5752– 5761 (2000).
Sarafianos, S.G. et al. Trapping HIV-1 reverse transcriptase before and after translocation on DNA. J. Biol. Chem. 278, 16280– 16288 (2003).
DeCorte, B.L. et al. Improved strategies for postoligomerization synthesis of oligodeoxynucleotides bearing structurally defined adducts at the N2 position of deoxyguanosine. Chem. Res. Toxicol. 9, 630– 637 (1996).
Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307– 326 (1997).
Brunger, A.T. et al. Crystallography & NMR system (CNS): a new software system for macromolecular structure determination. Acta Crystallogr. D 54, 905– 921 (1998).
Jones, T.A., Zou, J.Y., Cowan, S.W. & Kjeldgaard, M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47, 110– 119 (1991).
Acknowledgements
We thank the staffs at Advanced Photon Source BioCARS and CHESS for assistance with synchrotron data collection. We also thank other members of the Arnold lab including D. Sheng, K. Das, D. Oren and J. Birktoft for assistance and helpful discussions. We are grateful for support from US National Institutes of Health grants T32 AI50382 (National Research Service Award postdoctoral fellowship to S.T.), R01 AI27690 (MERIT award to E.A.) and P01 GM56690 (to E.A., S.S.H. and R.A.J.). S.H.H.'s laboratory has been supported by the US National Institute of General Medical Sciences and the US National Cancer Institute.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
During portions of this study, S.T. and E.A. were paid consultants for Gilead Sciences and L.K.N., K.W., M.D.M. and C.S.G. were employees of Gilead Sciences.
Supplementary information
Rights and permissions
About this article
Cite this article
Tuske, S., Sarafianos, S., Clark, A. et al. Structures of HIV-1 RT–DNA complexes before and after incorporation of the anti-AIDS drug tenofovir. Nat Struct Mol Biol 11, 469–474 (2004). https://doi.org/10.1038/nsmb760
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb760
This article is cited by
-
Structural features in common of HBV and HIV-1 resistance against chirally-distinct nucleoside analogues entecavir and lamivudine
Scientific Reports (2020)
-
Elucidating molecular interactions of L-nucleotides with HIV-1 reverse transcriptase and mechanism of M184V-caused drug resistance
Communications Biology (2019)
-
Modeling the functional state of the reverse transcriptase of hepatitis B virus and its application to probing drug-protein interaction
BMC Bioinformatics (2016)
-
Hypersusceptibility mechanism of Tenofovir-resistant HIV to EFdA
Retrovirology (2013)
-
Binding sensitivity of adefovir to the polymerase from different genotypes of HBV: molecular modeling, docking and dynamics simulation studies
Acta Pharmacologica Sinica (2013)