Abstract
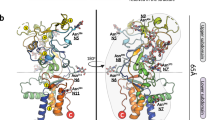
Although many viral receptors have been identified, the ways in which they interact with their cognate viruses are not understood at the molecular level. We have determined the X-ray structure of a complex between calcium-containing modules of the very low-density lipoprotein receptor and the minor group human rhinovirus HRV2. The receptor binds close to the icosahedral five-fold vertex, with only one module per virus protomer. The binding face of this module is defined by acidic calcium-chelating residues and, in particular, by an exposed tryptophan that is highly conserved. The attachment site on the virus involves only residues from VP1, particularly a lysine strictly conserved in all minor group HRVs. The disposition of the attached ligand-binding repeats around the five-fold axis, together with the proximity of the N- and C-terminal ends of adjacent modules, suggests that more than one repeat in a single receptor molecule might attach simultaneously.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Accession codes
References
Fry, E.E. et al. The structure and function of a foot-and-mouth disease virus–oligosaccharide receptor complex. EMBO J. 18, 543–554 (1999).
Greve, J.M. et al. The major human rhinovirus receptor is ICAM-1. Cell 56, 839–847 (1989).
Staunton, D.E. et al. A cell adhesion molecule, ICAM-1, is the major surface receptor for rhinoviruses. Cell 56, 849–853 (1989).
Hofer, F. et al. Members of the low density lipoprotein receptor family mediate cell entry of a minor-group common cold virus. Proc. Natl. Acad. Sci. USA 91, 1839–1842 (1994).
Marlovits, T.C., Abrahamsberg, C. & Blaas, D. Very-low-density lipoprotein receptor fragment shed from HeLa cells inhibits human rhinovirus infection. J. Virol. 72, 10246–10250 (1998).
Herz, J. & Marschang, P. Coaxing the LDL receptor family into the fold. Cell 112, 289–292 (2003).
Fass, D., Blacklow, S., Kim, P.S. & Berger, J.M. Molecular basis of familial hypercholesterolaemia from structure of LDL receptor module. Nature 388, 691–693 (1997).
Rudenko, G. et al. Structure of the LDL receptor extracellular domain at endosomal pH. Science 298, 2353–2358 (2002).
Davis, C.G. et al. Acid-dependent ligand dissociation and recycling of LDL receptor mediated by growth factor homology region. Nature 326, 760–765 (1987).
Nykjaer, A. & Willnow, T.E. The low-density lipoprotein receptor gene family: a cellular Swiss army knife? Trends Cell Biol. 12, 273–280 (2002).
Rossmann, M.G. et al. Structure of a human common cold virus and functional relationship to other picornaviruses. Nature 317, 145–153 (1985).
Badger, J. et al. Structural analysis of a series of antiviral agents complexed with human rhinovirus 14. Proc. Natl. Acad. Sci. USA 85, 3304–3308 (1988).
Hewat, E.A. et al. The cellular receptor to human rhinovirus 2 binds around the 5-fold axis and not in the canyon: a structural view. EMBO J. 19, 6317–6325 (2000).
Neumann, E., Moser, R., Snyers, L., Blaas, D. & Hewat, E.A. A cellular receptor of human rhinovirus type 2, the very-low-density lipoprotein receptor, binds to two neighboring proteins of the viral capsid. J. Virol. 77, 8504–8511 (2003).
Dolmer, K., Huang, W. & Gettins, P.G. NMR solution structure of complement-like repeat CR3 from the low density lipoprotein receptor-related protein. Evidence for specific binding to the receptor binding domain of human α(2)-macroglobulin. J. Biol. Chem. 275, 3264–3269 (2000).
Daly, N.L., Scanlon, M.J., Djordjevic, J.T., Kroon, P.A. & Smith, R. Three-dimensional structure of a cysteine-rich repeat from the low-density lipoprotein receptor. Proc. Natl. Acad. Sci. USA 92, 6334–6338 (1995).
Daly, N.L., Djordjevic, J.T., Kroon, P.A. & Smith, R. Three-dimensional structure of the second cysteine-rich repeat from the human low-density lipoprotein receptor. Biochemistry 34, 14474–14481 (1995).
Huang, W., Dolmer, K. & Gettins, P.G.W. NMR solution structure of complement-like repeat CR8 from the low density lipoprotein receptor-related protein. J. Biol. Chem. 274, 14130–14136 (1999).
Verdaguer, N., Blaas, D. & Fita, I. Structure of human rhinovirus serotype 2 (HRV2). J. Mol. Biol. 300, 1179–1194 (2000).
Rossmann, M.G. Viral cell recognition and entry. Protein Sci. 3, 1712–1725 (1994).
Lo Conte, L., Chothia, C. & Janin, J. The atomic structure of protein-protein recognition sites. J. Mol. Biol. 285, 2177–2198 (1999).
Fox, G. et al. The cell attachment site on foot-and-mouth disease virus includes the amino acid sequence RGD (arginine-glycine-aspartic acid). J. Gen. Virol. 70, 625–637 (1989).
Appleyard, G. et al. Neutralization epitopes of human rhinovirus type 2. J. Gen. Virol. 71, 1275–1282 (1990).
Vlasak, M., Blomqvist, S., Hovi, T., Hewat, E. & Blaas, D. Sequence and structure of human rhinoviruses reveal the basis of receptor discrimination. J. Virol. 77, 6923–6930 (2003).
Duechler, M., Ketter, S., Skern, T., Kuechler, E. & Blaas, D. Rhinoviral receptor discrimination: mutational changes in the canyon regions of human rhinovirus types 2 and 14 indicate a different site of interaction. J. Gen. Virol. 74, 2287–2291 (1993).
Brabec, M., Baravalle, G., Blaas, D. & Fuchs, R. Conformational changes, plasma membrane penetration, and infection by human rhinovirus type 2: role of receptors and low pH. J. Virol. 77, 5370–5377 (2003).
Oka, K. et al. Mouse very-low-density-lipoprotein receptor (VLDLR) cDNA cloning, tissue-specific expression and evolutionary relationship with the low-density-lipoprotein receptor. Eur. J. Biochem. 224, 975–982 (1994).
Reithmayer, M., Reischl, A., Snyers, L. & Blaas, D. Species-specific receptor recognition by a minor-group human rhinovirus (HRV): HRV serotype 1A distinguishes between the murine and the human low-density lipoprotein receptor. J. Virol. 76, 6957–6965 (2002).
Marlovits, T.C. et al. Recombinant soluble low density lipoprotein receptor fragment inhibits minor group rhinovirus infection in vitro . FASEB J. 12, 695–703 (1998).
Marlovits, T.C., Abrahamsberg, C. & Blaas, D. Soluble LDL minireceptors. Minimal structure requirements for recognition of minor group human rhinovirus. J. Biol. Chem. 273, 33835–33840 (1998).
Goldstein, J.L. & Brown, M.S. Binding and degradation of low density lipoproteins by cultured human fibroblasts. Comparison of cells from a normal subject and from a patient with homozygous familial hypercholesterolemia. J. Biol. Chem. 249, 5153–5162 (1974).
Ronacher, B., Marlovits, T.C., Moser, R. & Blaas, D. Expression and folding of human very-low-density lipoprotein receptor fragments: neutralization capacity toward human rhinovirus HRV2. Virology 278, 541–550 (2000).
Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997).
Brünger, A.T. X-PLOR: A System for X-ray Crystallography and NMR (Yale Univ. Press, New Haven, 1993).
Diprose, J.M. et al., translocation portals for the substrates and products of a viral transcription complex: the bluetonge virus core. EMBO J. 20, 7229–7239 (2001).
Roussel, A. & Cambillau, C. Turbo-Frodo. In Silicon Graphics Geometry Partners Directory 77–79 (Silicon Graphics, Mountain View, California, USA, 1989).
Jones, T.A., Zou, J.Y., Cowan, S.W. & Kjeldgaard . Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47, 110–119 (1991).
Brünger, A.T. et al. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D 54, 905–921 (1998).
Laskowski, R.A., MacArthur, M.W., Moss, D.S. & Thornton, J.M. PROCHECK: programs for checking the quality of protein structures. J. Appl. Cryst. 26, 283–291 (1993).
Herdy, B., Snyers, L., Reithmayer, M., Hinterdorfer, P. & Blaas, D. Identification of the HRV1A binding site on murine LDLR using human-mouse chimeras. J. Virol. (in the press).
Acknowledgements
This work was supported by grants BIO2002-00517 and BIO2002-04419 of Comision Interministerial de Ciencia y Tecnología to N.V and I.F, respectively, and by the Austrian Science Foundation, grant number P-14503, to D.B. Data were collected at the European Molecular Biology Laboratory protein crystallography beamlines ID14.4 and ID29 at European Synchrotron Radiation Facility (ESRF, Grenoble) within a Block Allocation Group (BAG Barcelona). Financial support was provided by the ESRF and by grant HPRI-CT-1999-00022 of the European Union. We thank I. Goesler for expert technical assistance, E. Hewat for sending us the electron microscopy maps before publication and M.G. Rossmann, T. Skern and P.C. Loewen for critically reading the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Verdaguer, N., Fita, I., Reithmayer, M. et al. X-ray structure of a minor group human rhinovirus bound to a fragment of its cellular receptor protein. Nat Struct Mol Biol 11, 429–434 (2004). https://doi.org/10.1038/nsmb753
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb753
This article is cited by
-
The low-density lipoprotein receptor promotes infection of multiple encephalitic alphaviruses
Nature Communications (2024)
-
Strong relationship between cholesterol, low-density lipoprotein receptor, Na+/H+ exchanger, and SARS-COV-2: this association may be the cause of death in the patient with COVID-19
Lipids in Health and Disease (2021)
-
Structure of Venezuelan equine encephalitis virus in complex with the LDLRAD3 receptor
Nature (2021)
-
Conjunctivitis as a Sentinel of SARS-CoV-2 Infection: a Need of Revision for Mild Symptoms
SN Comprehensive Clinical Medicine (2020)
-
Structural basis for the recognition of LDL-receptor family members by VSV glycoprotein
Nature Communications (2018)