Abstract
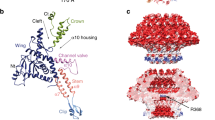
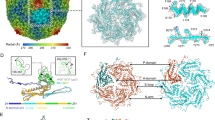
Bacteriophage P22 infects Salmonella enterica by injecting its genetic material through the cell envelope. During infection, a specialized tail needle, gp26, is injected into the host, likely piercing a hole in the host cell envelope. The 2.1-Å crystal structure of gp26 reveals a 240-Å elongated protein fiber formed by two trimeric coiled-coil domains interrupted by a triple β-helix. The N terminus of gp26 plugs the portal protein channel, retaining the genetic material inside the virion. The C-terminal tip of the fiber exposes β-hairpins with hydrophobic tips similar to those seen in class II fusion peptides. The α-helical core connecting these two functionally polarized tips presents four trimerization octads with consensus sequence IXXLXXXV. The slender conformation of the gp26 fiber minimizes the surface exposed to solvent, which is consistent with the idea that gp26 traverses the cell envelope lipid bilayers.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Accession codes
References
Lander, G.C. et al. The structure of an infectious p22 virion shows the signal for headful DNA packaging. Science 312, 1791–1795 (2006).
Chang, J., Weigele, P., King, J., Chiu, W. & Jiang, W. Cryo-EM asymmetric reconstruction of bacteriophage P22 reveals organization of its DNA packaging and infecting machinery. Structure 14, 1073–1082 (2006).
Strauss, H. & King, J. Steps in the stabilization of newly packaged DNA during phage P22 morphogenesis. J. Mol. Biol. 172, 523–543 (1984).
Lenk, E., Casjens, S., Weeks, J. & King, J. Intracellular visualization of precursor capsids in phage P22 mutant infected cells. Virology 68, 182–199 (1975).
Berget, P.B. & Poteete, A.R. Structure and functions of the bacteriophage P22 tail protein. J. Virol. 34, 234–243 (1980).
Molineux, I.J. No syringes please, ejection of phage T7 DNA from the virion is enzyme driven. Mol. Microbiol. 40, 1–8 (2001).
Israel, V. A model for the adsorption of phage P22 to Salmonella typhimurium. J. Gen. Virol. 40, 669–673 (1978).
Bhardwaj, A., Olia, A.S., Walker-Kopp, N. & Cingolani, G. Domain organization and polarity of tail needle GP26 in the portal vertex structure of bacteriophage P22. J. Mol. Biol. 371, 374–387 (2007).
Andrews, D. et al. Bacteriophage P22 tail accessory factor GP26 is a long triple-stranded coiled-coil. J. Biol. Chem. 280, 5929–5933 (2005).
Israel, V. E proteins of bacteriophage P22. I. Identification and ejection from wild-type and defective particles. J. Virol. 23, 91–97 (1977).
Moak, M. & Molineux, I.J. Peptidoglycan hydrolytic activities associated with bacteriophage virions. Mol. Microbiol. 51, 1169–1183 (2004).
Cingolani, G., Andrews, D. & Casjens, S. Crystallogenesis of bacteriophage P22 tail accessory factor gp26 at acidic and neutral pH. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 62, 477–482 (2006).
Bullough, P.A., Hughson, F.M., Skehel, J.J. & Wiley, D.C. Structure of influenza haemagglutinin at the pH of membrane fusion. Nature 371, 37–43 (1994).
Eckert, D.M. & Kim, P.S. Mechanisms of viral membrane fusion and its inhibition. Annu. Rev. Biochem. 70, 777–810 (2001).
Crick, F.H.C. The packing of α-helices: simple coiled-coils. Acta Crystallogr. 6, 689–697 (1953).
Burkhard, P., Kammerer, R.A., Steinmetz, M.O., Bourenkov, G.P. & Aebi, U. The coiled-coil trigger site of the rod domain of cortexillin I unveils a distinct network of interhelical and intrahelical salt bridges. Structure 8, 223–230 (2000).
Stummeyer, K., Dickmanns, A., Muhlenhoff, M., Gerardy-Schahn, R. & Ficner, R. Crystal structure of the polysialic acid-degrading endosialidase of bacteriophage K1F. Nat. Struct. Mol. Biol. 12, 90–96 (2005).
Tang, L., Marion, W.R., Cingolani, G., Prevelige, P.E. & Johnson, J.E. Three-dimensional structure of the bacteriophage P22 tail machine. EMBO J. 24, 2087–2095 (2005).
Modis, Y., Ogata, S., Clements, D. & Harrison, S.C. Structure of the dengue virus envelope protein after membrane fusion. Nature 427, 313–319 (2004).
Evilevitch, A., Lavelle, L., Knobler, C.M., Raspaud, E. & Gelbart, W.M. Osmotic pressure inhibition of DNA ejection from phage. Proc. Natl. Acad. Sci. USA 100, 9292–9295 (2003).
Kindt, J., Tzlil, S., Ben-Shaul, A. & Gelbart, W.M. DNA packaging and ejection forces in bacteriophage. Proc. Natl. Acad. Sci. USA 98, 13671–13674 (2001).
Coombs, D.H. & Ariska, F. in Molecular Biology of Bacteriophage T4 (ed. Karam, J.D.) 259–281 (American Society of Microbiology, Washington DC, 1994).
Goldberg, E., Grinius, L. & Letellier, L. in Molecular Biology of Bacteriophage T4 (ed. Karam, J.D.) 347–356 (American Society of Microbiology, Washington DC, 1994).
Kanamaru, S. et al. Structure of the cell-puncturing device of bacteriophage T4. Nature 415, 553–557 (2002).
Nakagawa, H., Arisaka, F. & Ishii, S. Isolation and characterization of the bacteriophage T4 tail-associated lysozyme. J. Virol. 54, 460–466 (1985).
Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997).
Collaborative Computational Project, Number 4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50, 760–763 (1994).
Terwilliger, T.C. SOLVE and RESOLVE: automated structure solution and density modification. Methods Enzymol. 374, 22–37 (2003).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004).
Brunger, A.T. et al. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54, 905–921 (1998).
Sheldrick, G.M. & Schneider, T.R. SHELXL: High-resolution refinement. Methods Enzymol. 277, 319–343 (1997).
Larsen, N.A. et al. Structure determination of a cocaine hydrolytic antibody from a pseudomerohedrally twinned crystal. Acta Crystallogr. D Biol. Crystallogr. 58, 2055–2059 (2002).
Yeates, T.O. & Fam, B.C. Protein crystals and their evil twins. Structure 7, R25–R29 (1999).
Pettersen, E.F. et al. UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Merritt, E.A. & Murphy, M.E. Raster3D Version 2.0. A program for photorealistic molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 50, 869–873 (1994).
Acknowledgements
We thank N. Walker for technical help. We are grateful to V. Stojanoff at the NSLS and to the macCHESS staff for beam time and assistance in data collection. This work was supported in part by US National Science Foundation grant MCB-990526 to S.C.
Author information
Authors and Affiliations
Contributions
A.S.O. and G.C. contributed to the structural and biochemical studies. S.C. participated in the design and coordination of the study. A.S.O., S.C. and G.C. wrote the manuscript.
Corresponding author
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–3 and Supplementary Table 1 (PDF 4534 kb)
Rights and permissions
About this article
Cite this article
Olia, A., Casjens, S. & Cingolani, G. Structure of phage P22 cell envelope–penetrating needle. Nat Struct Mol Biol 14, 1221–1226 (2007). https://doi.org/10.1038/nsmb1317
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb1317
This article is cited by
-
Structure and proposed DNA delivery mechanism of a marine roseophage
Nature Communications (2023)
-
Targeting mechanisms of tailed bacteriophages
Nature Reviews Microbiology (2018)
-
Artificial bio-nanomachines based on protein needles derived from bacteriophage T4
Biophysical Reviews (2018)
-
Portal protein functions akin to a DNA-sensor that couples genome-packaging to icosahedral capsid maturation
Nature Communications (2017)