Abstract
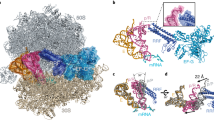
In bacteria, disassembly of the ribosome at the end of translation is facilitated by an essential protein factor termed ribosome recycling factor (RRF), which works in concert with elongation factor G. Here we describe the crystal structure of the Thermus thermophilus RRF bound to a 70S ribosomal complex containing a stop codon in the A site, a transfer RNA anticodon stem-loop in the P site and tRNAfMet in the E site. The work demonstrates that structures of translation factors bound to 70S ribosomes can be determined at reasonably high resolution. Contrary to earlier reports, we did not observe any RRF-induced changes in bridges connecting the two subunits. This suggests that such changes are not a direct requirement for or consequence of RRF binding but possibly arise from the subsequent stabilization of a hybrid state of the ribosome.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Hirashima, A. & Kaji, A. Purification and properties of ribosome-releasing factor. Biochemistry 11, 4037–4044 (1972).
Janosi, L., Shimizu, I. & Kaji, A. Ribosome recycling factor (ribosome releasing factor) is essential for bacterial growth. Proc. Natl. Acad. Sci. USA 91, 4249–4253 (1994).
Hirashima, A. & Kaji, A. Role of elongation factor G and a protein factor on the release of ribosomes from messenger ribonucleic acid. J. Biol. Chem. 248, 7580–7587 (1973).
Selmer, M., Al-Karadaghi, S., Hirokawa, G., Kaji, A. & Liljas, A. Crystal structure of Thermotoga maritima ribosome recycling factor: a tRNA mimic. Science 286, 2349–2352 (1999).
Kim, K.K., Min, K. & Suh, S.W. Crystal structure of the ribosome recycling factor from Escherichia coli. EMBO J. 19, 2362–2370 (2000).
Toyoda, T. et al. Crystal structure combined with genetic analysis of the Thermus thermophilus ribosome recycling factor shows that a flexible hinge may act as a functional switch. RNA 6, 1432–1444 (2000).
Nakano, H. et al. Structure and binding mode of a ribosome recycling factor (RRF) from mesophilic bacterium. J. Biol. Chem. 278, 3427–3436 (2003).
Yoshida, T. et al. Solution structure of the ribosome recycling factor from Aquifex aeolicus. Biochemistry 40, 2387–2396 (2001).
Yoshida, T. et al. Characteristic domain motion in the ribosome recycling factor revealed by 15N NMR relaxation experiments and molecular dynamics simulations. Biochemistry 42, 4101–4107 (2003).
Lancaster, L., Kiel, M.C., Kaji, A. & Noller, H.F. Orientation of ribosome recycling factor in the ribosome from directed hydroxyl radical probing. Cell 111, 129–140 (2002).
Agrawal, R.K. et al. Visualization of ribosome-recycling factor on the Escherichia coli 70S ribosome: functional implications. Proc. Natl. Acad. Sci. USA 101, 8900–8905 (2004).
Gao, N. et al. Mechanism for the disassembly of the posttermination complex inferred from cryo-EM studies. Mol. Cell 18, 663–674 (2005).
Wilson, D.N. et al. X-ray crystallography study on ribosome recycling: the mechanism of binding and action of RRF on the 50S ribosomal subunit. EMBO J. 24, 251–260 (2005).
Rao, A.R. & Varshney, U. Specific interaction between the ribosome recycling factor and the elongation factor G from Mycobacterium tuberculosis mediates peptidyl-tRNA release and ribosome recycling in Escherichia coli. EMBO J. 20, 2977–2986 (2001).
Ito, K., Fujiwara, T., Toyoda, T. & Nakamura, Y. Elongation factor G participates in ribosome disassembly by interacting with ribosome recycling factor at their tRNA-mimicry domains. Mol. Cell 9, 1263–1272 (2002).
Guo, P., Zhang, L., Zhang, H., Feng, Y. & Jing, G. Domain II plays a crucial role in the function of ribosome recycling factor. Biochem. J. 393, 767–777 (2006).
Hirashima, A. & Kaji, A. Factor-dependent release of ribosomes from messenger RNA. Requirement for two heat-stable factors. J. Mol. Biol. 65, 43–58 (1972).
Karimi, R., Pavlov, M.Y., Buckingham, R.H. & Ehrenberg, M. Novel roles for classical factors at the interface between translation termination and initiation. Mol. Cell 3, 601–609 (1999).
Hirokawa, G. et al. Post-termination complex disassembly by ribosome recycling factor, a functional tRNA mimic. EMBO J. 21, 2272–2281 (2002).
Peske, F., Rodnina, M.V. & Wintermeyer, W. Sequence of steps in ribosome recycling as defined by kinetic analysis. Mol. Cell 18, 403–412 (2005).
Selmer, M. et al. Structure of the 70S ribosome complexed with mRNA and tRNA. Science 313, 1935–1942 (2006).
Maguire, B.A., Beniaminov, A.D., Ramu, H., Mankin, A.S. & Zimmermann, R.A. A protein component at the heart of an RNA machine: the importance of protein l27 for the function of the bacterial ribosome. Mol. Cell 20, 427–435 (2005).
Yusupov, M.M. et al. Crystal structure of the ribosome at 5.5 A resolution. Science 292, 883–896 (2001).
Schuwirth, B.S. et al. Structures of the bacterial ribosome at 3.5 A resolution. Science 310, 827–834 (2005).
Korostelev, A., Trakhanov, S., Laurberg, M. & Noller, H.F. Crystal structure of a 70S ribosome-tRNA complex reveals functional interactions and rearrangements. Cell 126, 1065–1077 (2006).
Ermolenko, D.N. et al. The antibiotic viomycin traps the ribosome in an intermediate state of translocation. Nat. Struct. Mol. Biol. 14, 493–497 (2007).
Frank, J. & Agrawal, R.K. A ratchet-like inter-subunit reorganization of the ribosome during translocation. Nature 406, 318–322 (2000).
Petry, S. et al. Crystal structures of the ribosome in complex with release factors RF1 and RF2 bound to a cognate stop codon. Cell 123, 1255–1266 (2005).
Schmitt, E., Panvert, M., Blanquet, S. & Mechulam, Y. Crystal structure of methionyl-tRNAfMet transformylase complexed with the initiator formyl-methionyl-tRNAfMet. EMBO J. 17, 6819–6826 (1998).
Kabsch, W. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J. Appl. Cryst. 26, 795–800 (1993).
Brünger, A.T. et al. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54, 905–921 (1998).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004).
Acknowledgements
We thank F. Murphy for help with data collection during optimization of crystallization conditions, M. MacDonald for help with screening at beamline 14.1 at the Synchrotron Radiation Source, and R. Ravelli, J. McCarthy and G. Leonard at beamlines ID14-1, ID14-3 and ID14-4 at the ESRF for help with screening and data collection. This work was supported by the Medical Research Council (UK), the US National Institutes of Health and the Agouron Institute, and fellowships from the Austrian Academy of Sciences (A.W.), the Boehringer Ingelheim Fonds (S.P.), the American Cancer Society (C.M.D.) and the Wenner-Gren foundation (M.S.).
Author information
Authors and Affiliations
Contributions
A.W. crystallized the complex with RRF, collected and analyzed the data and wrote the paper. S.P. and C.M.D. helped with optimization of mRNA constructs, crystallization and cryoprotection conditions, helped with data collection, initial processing and model building, and gave feedback on the manuscript. M.S. cloned, expressed and purified T. thermophilus RRF, suggested crystallization of its complex with the ribosome and provided feedback on the manuscript. A.C.K. purified ribosomes and tRNA. V.R. oversaw the work and helped with writing the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–3. (PDF 228 kb)
Rights and permissions
About this article
Cite this article
Weixlbaumer, A., Petry, S., Dunham, C. et al. Crystal structure of the ribosome recycling factor bound to the ribosome. Nat Struct Mol Biol 14, 733–737 (2007). https://doi.org/10.1038/nsmb1282
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb1282
This article is cited by
-
Distinct mechanisms of the human mitoribosome recycling and antibiotic resistance
Nature Communications (2021)
-
Structural basis for ribosome recycling by RRF and tRNA
Nature Structural & Molecular Biology (2020)
-
Small methyltransferase RlmH assembles a composite active site to methylate a ribosomal pseudouridine
Scientific Reports (2017)
-
HflX is a ribosome-splitting factor rescuing stalled ribosomes under stress conditions
Nature Structural & Molecular Biology (2015)
-
Mechanism of eIF6 release from the nascent 60S ribosomal subunit
Nature Structural & Molecular Biology (2015)