Abstract
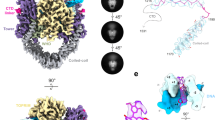
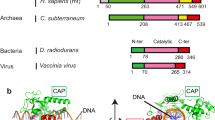
Type II topoisomerases help disentangle chromosomes to facilitate cell division. To advance understanding of the structure and dynamics of these essential enzymes, we have determined the crystal structure of an archaeal type IIB topoisomerase, topo VI, at 4.0-Å resolution. The 220-kDa heterotetramer adopts a 'twin-gate' architecture, in which a pair of ATPase domains at one end of the enzyme is poised to coordinate DNA movements into the enzyme and through a set of DNA-cleaving domains at the other end. Small-angle X-ray scattering studies show that nucleotide binding elicits a major structural reorganization that is propagated to the enzyme's DNA-cleavage center, explaining how ATP is coupled to DNA capture and strand scission. These data afford important insights into the mechanisms of topo VI and related proteins, including type IIA topoisomerases and the Spo11 meiotic recombination endonuclease.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Accession codes
References
Champoux, J.J. DNA topoisomerases: structure, function, and mechanism. Annu. Rev. Biochem. 70, 369–413 (2001).
Wang, J.C. Cellular roles of topoisomerases: a molecular perspective. Nat. Rev. Mol. Cell Biol. 3, 430–440 (2002).
Wang, J.C. Moving one DNA double helix through another by a type II DNA topoisomerase: the story of a simple molecular machine. Q. Rev. Biophys. 31, 107–144 (1998).
Corbett, K.D. & Berger, J.M. Structure, molecular mechanisms, and evolutionary relationships in DNA topoisomerases. Annu. Rev. Biophys. Biomol. Struct. 33, 95–118 (2004).
Gadelle, D., Filee, J., Buhler, C. & Forterre, P. Phylogenomics of type II DNA topoisomerases. Bioessays 25, 232–242 (2003).
Bergerat, A. et al. An atypical topoisomerase II from archaea with implications for meiotic recombination. Nature 386, 414–417 (1997).
Keeney, S., Giroux, C.N. & Kleckner, N. Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell 88, 375–384 (1997).
Nichols, M.D., DeAngelis, K., Keck, J.L. & Berger, J.M. Structure and function of an archaeal topoisomerase VI subunit with homology to the meiotic recombination factor Spo11. EMBO J. 18, 6177–6188 (1999).
Aravind, L., Leipe, D.D. & Koonin, E.V. Toprim: A conserved catalytic domain in type IA and II topoisomerases, DnaG-type primases, OLD family nucleases and RecR proteins. Nucleic Acids Res. 26, 4205–4213 (1998).
Dutta, R. & Inouye, M. GHKL, an emergent ATPase/kinase superfamily. Trends Biochem. Sci. 25, 24–28 (2000).
Wigley, D.B., Davies, G.J., Dodson, E.J., Maxwell, A. & Dodson, G. Crystal structure of an amino-terminal fragment of the DNA gyrase B protein. Nature 351, 624–629 (1991).
Corbett, K.D. & Berger, J.M. Structural dissection of ATP turnover in the prototypical GHL ATPase topo VI. Structure 13, 873–882 (2005).
Corbett, K.D. & Berger, J.M. Structure of the topoisomerase VI B subunit: implications for type II topoisomerase mechanism and evolution. EMBO J. 22, 151–163 (2003).
Roca, J. & Wang, J.C. The capture of a DNA double helix by an ATP-dependent protein clamp: a key step in DNA transport by type II DNA topoisomerases. Cell 71, 833–840 (1992).
Buhler, C., Lebbink, J.H.G., Bocs, C., Ladenstein, R. & Forterre, P. DNA topoisomerase VI generates ATP-dependent double-strand breaks with two-nucleotide overhangs. J. Biol. Chem. 276, 37215–37222 (2001).
Baird, C.L., Harkins, T.T., Morris, S.K. & Lindsley, J.E. Topoisomerase II drives DNA transport by hydrolyzing one ATP. Proc. Natl. Acad. Sci. USA 96, 13685–13690 (1999).
Roca, J. & Wang, J.C. DNA transport by a type II DNA topoisomerase: evidence in favor of a two-gate mechanism. Cell 77, 609–616 (1994).
Berger, J.M., Gamblin, S.J., Harrison, S.C. & Wang, J.C. Structure and mechanism of DNA topoisomerase II. Nature 379, 225–232 (1996).
Bork, P., Holm, L. & Sander, C. The immunoglobulin fold. Structural classification, sequence patterns and common core. J. Mol. Biol. 242, 309–320 (1994).
Xu, G.Y. et al. Solution structure of a cellulose-binding domain from Cellulomonas fimi by nuclear magnetic resonance spectroscopy. Biochemistry 34, 6993–7009 (1995).
Jenner, L., Husted, L., Thirup, S., Sottrup-Jensen, L. & Nyborg, J. Crystal structure of the receptor-binding domain of a 2-macroglobulin. Structure 6, 595–604 (1998).
Reece, R.J. & Maxwell, A. The C-terminal domain of the Escherichia coli DNA gyrase A subunit is a DNA-binding protein. Nucleic Acids Res. 19, 1399–1405 (1991).
Corbett, K.D., Schoeffler, A.J., Thomsen, N.D. & Berger, J.M. The structural basis for substrate specificity in DNA topoisomerase IV. J. Mol. Biol. 351, 545–561 (2005).
Corbett, K.D., Shultzaberger, R.K. & Berger, J.M. The C-terminal domain of DNA gyrase A adopts a DNA-bending β-pinwheel fold. Proc. Natl. Acad. Sci. USA 101, 7293–7298 (2004).
Ward, D. & Newton, A. Requirement of topoisomerase IV parC and parE genes for cell cycle progression and developmental regulation in Caulobacter crescentus. Mol. Microbiol. 26, 897–910 (1997).
Crisona, N.J., Strick, T.R., Bensimon, D., Croquette, V. & Cozzarelli, N.R. Preferential relaxation of positively supercoiled DNA by E. coli topoisomerase IV in single-molecule and ensemble measurements. Genes Dev. 14, 2881–2892 (2000).
Peng, H. & Marians, K.J. The interaction of Escherichia coli topoisomerase IV with DNA. J. Biol. Chem. 270, 25286–25290 (1995).
Wang, S.C. & Shapiro, L. The topoisomerase IV ParC subunit colocalizes with the Caulobacter replisome and is required for polar localization of replication origins. Proc. Natl. Acad. Sci. USA 101, 9251–9256 (2004).
Espeli, O., Lee, C. & Marians, K.J. A physical and functional interaction between Escherichia coli FtsK and topoisomerase IV. J. Biol. Chem. 278, 44639–44644 (2003).
Svergun, D.I. & Koch, M.H.J. Small-angle scattering studies of biological macromolecules in solution. Rep. Prog. Phys. 66, 1735–1782 (2003).
Svergun, D.I. Determination of the regularization parameter in indirect-transform methods using perceptual criteria. J. Appl. Cryst. 25, 495–503 (1992).
Svergun, D.I. Restoring low resolution structure of biological macromolecules from solution scattering using simulated annealing. Biophys. J. 76, 2879–2886 (1999).
Kozin, M.B. & Svergun, D.I. Automated matching of high- and low-resolution structural models. J. Appl. Cryst. 34, 33–41 (2001).
Svergun, D.I., Barberato, C. & Koch, M.H.J. CRYSOL - a program to evaluate x-ray solution scattering of biological macromolecules from atomic coordinates. J. Appl. Cryst. 28, 768–773 (1995).
Konarev, P.V., Volkov, V.V., Sokolova, A.V., Koch, M.H.J. & Svergun, D.I. PRIMUS: a Windows PC-based system for small-angle scattering data analysis. J. Appl. Cryst. 36, 1277–1282 (2003).
Buhler, C., Gadelle, D., Forterre, P., Wang, J.C. & Bergerat, A. Reconstitution of DNA topoisomerase VI of the thermophilic archaeon Sulfolobus shibatae from subunits separately overexpressed in Escherichia coli. Nucleic Acids Res. 26, 5157–5162 (1998).
Morais Cabral, J.H. et al. Crystal structure of the breakage-reunion domain of DNA gyrase. Nature 388, 903–906 (1997).
Tingey, A.P. & Maxwell, A. Probing the role of the ATP-operated clamp in the strand-passage reaction of DNA gyrase. Nucleic Acids Res. 24, 4868–4873 (1996).
Williams, N.L., Howells, A.J. & Maxwell, A. Locking the ATP-operated clamp of DNA gyrase: probing the mechanism of strand passage. J. Mol. Biol. 306, 969–984 (2001).
Lamour, V., Hoermann, L., Jeltsch, J-M., Oudet, P. & Moras, D. An open conformation of the Thermus thermophilus gyrase B ATP-binding domain. J. Biol. Chem. 277, 18947–18953 (2002).
Wei, H., Ruthenburg, A.J., Bechis, S.K. & Verdine, G.L. Nucleotide-dependent domain movement in the ATPase domain of a human type IIA DNA topoisomerase. J. Biol. Chem. 280, 37041–37047 (2005).
Morrison, A. & Cozzarelli, N.R. Site-specific cleavage of DNA by E. coli DNA gyrase. Cell 17, 175–184 (1979).
Sander, M. & Hsieh, T. Double strand DNA cleavage by type II DNA topoisomerase from Drosophila melanogaster. J. Biol. Chem. 258, 8421–8428 (1983).
Roca, J. The path of the DNA along the dimer interface of topoisomerase II. J. Biol. Chem. 279, 25783–25788 (2004).
Trigueros, S., Salceda, J., Bermudez, I., Fernandez, X. & Roca, J. Asymmetric removal of supercoils suggests how topoisomerase II simplifies DNA topology. J. Mol. Biol. 335, 723–731 (2004).
Rybenkov, V.V., Ullsperger, C., Vologodskii, A.V. & Cozzarelli, N.R. Simplification of DNA topology below equilibrium values by type II topoisomerases. Science 277, 690–693 (1997).
Keeney, S. & Neale, M.J. Initiation of meiotic recombination by formation of DNA double-strand breaks: mechanism and regulation. Biochem. Soc. Trans. 34, 523–525 (2006).
Nag, D.K., Pata, J.D., Sironi, M., Flood, D.R. & Hart, A.M. Both conserved and non-conserved regions of Spo11 are essential for meiotic recombination initiation in yeast. Mol. Genet. Genomics 276, 313–321 (2006).
Tan, S. A modular polycistronic expression system for overexpressing protein complexes in Escherichia coli. Protein Expr. Purif. 21, 224–234 (2001).
Kapust, R.B. & Waugh, D.S. Escherichia coli maltose-binding protein is uncommonly effective at promoting the solubility of polypeptides to which it is fused. Protein Sci. 8, 1668–1674 (1999).
MacDowell, A.A. et al. Suite of three protein crystallography beamlines with single superconducting bend magnet as the source. J. Synchrotron Radiat. 11, 447–455 (2004).
Benedetti, P., Silvestri, A., Fiorani, P. & Wang, J.C. Study of yeast DNA topoisomerase II and its truncation derivatives by transmission electron microscopy. J. Biol. Chem. 272, 12132–12137 (1997).
Kirchhausen, T., Wang, J.C. & Harrison, S.C. DNA gyrase and its complexes with DNA: direct observation by electron microscopy. Cell 41, 933–943 (1985).
Tyler, J.M. & Branton, D. Rotary shadowing of extended molecules dried from glycerol. J. Ultrastruct. Res. 71, 95–102 (1980).
Acknowledgements
This work is dedicated to the memory of Nicholas R. Cozzarelli, who provided invaluable advice and criticism to both K.D.C. and J.M.B. over many years. The authors thank J. Holton, G. Meigs and J. Tanamachi for assistance at ALS beamline 8.3.1, G. Hura and D. S. Classen for assistance at ALS beamline 12.3.1, A. Bergerat and J. Wang (Harvard University) for the gift of S. shibatae topo VI subunits, M. Nollmann for advice on SAXS methodology, and members of the Berger laboratory for helpful advice. J.M.B. acknowledges support from the US National Cancer Institute (CA077373), and P.B. acknowledges support from the Ministero dell'Instruzione, dell'Università e della Ricerca Cofinanziamento, Fondo Investimenti Ricerca di Base, Ministero della Salute and Genomica Funzionale Consiglio Nazionale delle Ricerche.
Author information
Authors and Affiliations
Contributions
K.D.C. performed all experiments, with the exception of the EM, which was performed by P.B. K.D.C. and J.M.B. devised all experiments and prepared the manuscript together, with editorial assistance from P.B.
Note: Supplementary information is available on the Nature Structural & Molecular Biology website.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Fig. 1
Biochemical characterization of M. mazei topo VI. (PDF 620 kb)
Supplementary Fig. 2
Experimental and refined electron density maps. (PDF 1560 kb)
Supplementary Fig. 3
Sequence alignments. (PDF 208 kb)
Supplementary Figs. 4
Cross-linking of SsT6S424C. (PDF 597 kb)
Supplementary Fig. 5
Nucleotide addition to SsT6 results in a mixture of conformational states. (PDF 87 kb)
Supplementary Fig. 6
DAMMIN models of ATP-SsT6S424C. (PDF 1122 kb)
Supplementary Fig. 7
Manual modeling of ATP-SsT6S424C. (PDF 630 kb)
Supplementary Fig. 8
MmT6 does not simplify DNA topology below thermodynamic equilibrium. (PDF 250 kb)
Supplementary Video 1
Different conformational states of topo VI and proposed strand passage mechanism. The movie begins with the partially closed conformation of MmT6 observed in the crystal structure, transitions to the open state observed in the apo-SsT6 SAXS sample, then transitions to the fully-closed state observed in the ATP-SsT6S424C SAXS sample and also based on the structure of the nucleotide-mediated S. shibatae topo VI B-subunit dimer1. Subsequently, a proposed strand passage reaction is modeled based on these states and on the nucleotide-mediated transducer domain motion observed in previous structures of the S. shibatae topo VI B-subunit2. In the movie, the B-subunit GHKL and H2TH domains are colored yellow, the transducer domain orange, the A-subunit CAP domains green, Toprim domains blue, and G- and T-segment DNAs magenta and cyan, respectively. (MOV 6222 kb)
Rights and permissions
About this article
Cite this article
Corbett, K., Benedetti, P. & Berger, J. Holoenzyme assembly and ATP-mediated conformational dynamics of topoisomerase VI. Nat Struct Mol Biol 14, 611–619 (2007). https://doi.org/10.1038/nsmb1264
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb1264
This article is cited by
-
Structural and functional characterization of the Spo11 core complex
Nature Structural & Molecular Biology (2021)
-
A ubiquitin-like domain is required for stabilizing the N-terminal ATPase module of human SMCHD1
Communications Biology (2019)
-
Functional characterization of the meiosis-specific DNA double-strand break inducing factor SPO-11 from C. elegans
Scientific Reports (2017)
-
DNA G-segment bending is not the sole determinant of topology simplification by type II DNA topoisomerases
Scientific Reports (2014)