Abstract
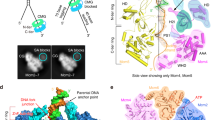
To reveal the mechanism of processive strand separation by superfamily-2 (SF2) 3′→5′ helicases, we determined apo and DNA-bound crystal structures of archaeal Hel308, a helicase that unwinds lagging strands and is related to human DNA polymerase θ. Our structure captures the duplex-unwinding reaction, shows that initial strand separation does not require ATP and identifies a prominent β-hairpin loop as the unwinding element. Similar loops in hepatitis C virus NS3 helicase and RNA-decay factors support the idea that this duplex-unwinding mechanism is applicable to a broad subset of SF2 helicases. Comparison with ATP-bound SF2 enzymes suggests that ATP promotes processive unwinding of 1 base pair by ratchet-like transport of the 3′ product strand. Our results provide a first structural framework for strand separation by processive SF2 3′→5′ helicases and reveal important mechanistic differences from SF1 helicases.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Linder, P. & Kowalczykowski, S. Helicases and NTP-driven nucleic acid machines: structure, function and roles in human disease. Nucleic Acids Res. 34, 4081 (2006).
Houseley, J., LaCava, J. & Tollervey, D. RNA-quality control by the exosome. Nat. Rev. Mol. Cell Biol. 7, 529–539 (2006).
Frick, D.N. Step-by-step progress toward understanding the hepatitis C virus RNA helicase. Hepatology 43, 1392–1395 (2006).
Gorbalenya, A.E., Koonin, E.V., Donchenko, A.P. & Blinov, V.M. Two related superfamilies of putative helicases involved in replication, recombination, repair and expression of DNA and RNA genomes. Nucleic Acids Res. 17, 4713–4730 (1989).
Velankar, S.S., Soultanas, P., Dillingham, M.S., Subramanya, H.S. & Wigley, D.B. Crystal structures of complexes of PcrA DNA helicase with a DNA substrate indicate an inchworm mechanism. Cell 97, 75–84 (1999).
Korolev, S., Hsieh, J., Gauss, G.H., Lohman, T.M. & Waksman, G. Major domain swiveling revealed by the crystal structures of complexes of E. coli Rep helicase bound to single-stranded DNA and ADP. Cell 90, 635–647 (1997).
Lee, J.Y., Yang, W. & Uvr, D. Helicase unwinds DNA one base pair at a time by a two-part power stroke. Cell 127, 1349–1360 (2006).
Fischer, C.J., Maluf, N.K. & Lohman, T.M. Mechanism of ATP-dependent translocation of E. coli UvrD monomers along single-stranded DNA. J. Mol. Biol. 344, 1287–1309 (2004).
Bennett, R.J. & Keck, J.L. Structure and function of RecQ DNA helicases. Crit. Rev. Biochem. Mol. Biol. 39, 79–97 (2004).
Seki, M., Marini, F. & Wood, R.D. POLQ (Pol theta), a DNA polymerase and DNA-dependent ATPase in human cells. Nucleic Acids Res. 31, 6117–6126 (2003).
Oshige, M., Aoyagi, N., Harris, P.V., Burtis, K.C. & Sakaguchi, K. A new DNA polymerase species from Drosophila melanogaster: a probable mus308 gene product. Mutat. Res. 433, 183–192 (1999).
Dumont, S. et al. RNA translocation and unwinding mechanism of HCV NS3 helicase and its coordination by ATP. Nature 439, 105–108 (2006).
Levin, M.K., Gurjar, M. & Patel, S.S. A Brownian motor mechanism of translocation and strand separation by hepatitis C virus helicase. Nat. Struct. Mol. Biol. 12, 429–435 (2005).
Serebrov, V. & Pyle, A.M. Periodic cycles of RNA unwinding and pausing by hepatitis C virus NS3 helicase. Nature 430, 476–480 (2004).
Sengoku, T., Nureki, O., Nakamura, A., Kobayashi, S. & Yokoyama, S. Structural basis for RNA unwinding by the DEAD-box protein Drosophila Vasa. Cell 125, 287–300 (2006).
Singleton, M.R., Scaife, S. & Wigley, D.B. Structural analysis of DNA replication fork reversal by RecG. Cell 107, 79–89 (2001).
Durr, H., Korner, C., Muller, M., Hickmann, V. & Hopfner, K.P. X-ray structures of the Sulfolobus solfataricus SWI2/SNF2 ATPase core and its complex with DNA. Cell 121, 363–373 (2005).
Marini, F. & Wood, R.D. A human DNA helicase homologous to the DNA cross-link sensitivity protein Mus308. J. Biol. Chem. 277, 8716–8723 (2002).
McCaffrey, R., St Johnston, D. & Gonzalez-Reyes, A. Drosophila mus301/spindle-C encodes a helicase with an essential role in double-strand DNA break repair and meiotic progression. Genetics 174, 1273–1285 (2006).
Yoshimura, M. et al. Vertebrate POLQ and POLbeta cooperate in base excision repair of oxidative DNA damage. Mol. Cell 24, 115–125 (2006).
Koonin, E.V., Wolf, Y.I. & Aravind, L. Prediction of the archaeal exosome and its connections with the proteasome and the translation and transcription machineries by a comparative-genomic approach. Genome Res. 11, 240–252 (2001).
Guy, C.P. & Bolt, E.L. Archaeal Hel308 helicase targets replication forks in vivo and in vitro and unwinds lagging strands. Nucleic Acids Res. 33, 3678–3690 (2005).
Fujikane, R., Komori, K., Shinagawa, H. & Ishino, Y. Identification of a novel helicase activity unwinding branched DNAs from the hyperthermophilic archaeon, Pyrococcus furiosus. J. Biol. Chem. 280, 12351–12358 (2005).
Singleton, M.R., Dillingham, M.S., Gaudier, M., Kowalczykowski, S.C. & Wigley, D.B. Crystal structure of RecBCD enzyme reveals a machine for processing DNA breaks. Nature 432, 187–193 (2004).
Bono, F., Ebert, J., Lorentzen, E. & Conti, E. The crystal structure of the exon junction complex reveals how it maintains a stable grip on mRNA. Cell 126, 713–725 (2006).
Andersen, C.B. et al. Structure of the exon junction core complex with a trapped DEAD-box ATPase bound to RNA. Science 313, 1968–1972 (2006).
Kim, J.L. et al. Hepatitis C virus NS3 RNA helicase domain with a bound oligonucleotide: the crystal structure provides insights into the mode of unwinding. Structure 6, 89–100 (1998).
Levin, M.K., Gurjar, M.M. & Patel, S.S. ATP binding modulates the nucleic acid affinity of hepatitis C virus helicase. J. Biol. Chem. 278, 23311–23316 (2003).
Lam, A.M., Keeney, D. & Frick, D.N. Two novel conserved motifs in the hepatitis C virus NS3 protein critical for helicase action. J. Biol. Chem. 278, 44514–44524 (2003).
Jankowsky, E., Gross, C.H., Shuman, S. & Pyle, A.M. Active disruption of an RNA-protein interaction by a DExH/D RNA helicase. Science 291, 121–125 (2001).
Kabsch, W. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J. Appl. Cryst. 26, 795–800 (1993).
Schneider, T.R. & Sheldrick, G.M. Substructure solution with SHELXD. Acta Crystallogr. D Biol. Crystallogr. 58, 1772–1779 (2002).
Turk, D. Weiterentwicklung eines programms fuer molekuelgraphik und elektrondichte-manipulation und seine anwendung auf verschiedene protein-strukturaufklaerungen. Thesis, Technical Univ. Munich, (1992).
Brunger, A.T. et al. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54, 905–921 (1998).
Collaborative Computational Project, Number 4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50, 760–763 (1994).
Acknowledgements
We thank S. Cui for comments on the manuscript, F. Uhlmann for collaborative efforts and scientific discussions, and the staff of the Swiss Light Source (beamline PXI) and the European Synchrotron Radiation Facility for generous beamtime allocation and help with data collection. K.-P.H. acknowledges grant support from the German research council (Deutsche Forschungsgemeinschaft HO2489/3-1 and SFB455), Human Frontiers of Science and the European Commission (integrated project 'DNA Repair').
Author information
Authors and Affiliations
Contributions
K.B. crystallized DNA-bound Hel308, determined structures and designed research; S.N. cloned the gene for A. fulgidus Hel308 and crystallized apo-Hel308; K.-P.H. designed research, participated in model building and evaluation, and wrote manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Fig. 1
Sequence alignment of A. fulgidus Hel308 with selected SF2/Ski2 family helicases. (PDF 629 kb)
Supplementary Fig. 2
Composit simulated annealed omit electron density shows how strands of a DNA duplex are separated by archaeal Hel208. (PDF 749 kb)
Supplementary Fig. 3
Comparison of apo- and DNA-bound archaeal Hel308. (PDF 688 kb)
Supplementary Fig. 4
Test for large-scale DNA- and ATP-induced conformational changes in archaeal Hel308 by limited proteolysis digestion using trypsin and chymotrypsin. (PDF 339 kb)
Rights and permissions
About this article
Cite this article
Büttner, K., Nehring, S. & Hopfner, KP. Structural basis for DNA duplex separation by a superfamily-2 helicase. Nat Struct Mol Biol 14, 647–652 (2007). https://doi.org/10.1038/nsmb1246
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb1246
This article is cited by
-
Energy-driven genome regulation by ATP-dependent chromatin remodellers
Nature Reviews Molecular Cell Biology (2024)
-
Genotype–phenotype correlations and novel molecular insights into the DHX30-associated neurodevelopmental disorders
Genome Medicine (2021)
-
Quaternary structural diversity in eukaryotic DNA polymerases: monomeric to multimeric form
Current Genetics (2020)
-
Structure and function relationships in mammalian DNA polymerases
Cellular and Molecular Life Sciences (2020)
-
Molecular basis of microhomology-mediated end-joining by purified full-length Polθ
Nature Communications (2019)