Abstract
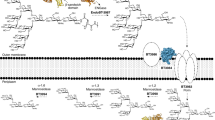
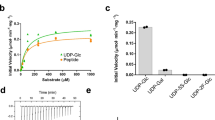
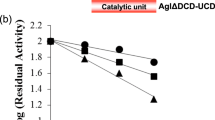
O-GlcNAc is an abundant post-translational modification of serine and threonine residues of nucleocytoplasmic proteins. This modification, found only within higher eukaryotes, is a dynamic modification that is often reciprocal to phosphorylation. In a manner analogous to phosphatases, a glycoside hydrolase termed O-GlcNAcase cleaves O-GlcNAc from modified proteins. Enzymes with high sequence similarity to human O-GlcNAcase are also found in human pathogens and symbionts. We report the three-dimensional structure of O-GlcNAcase from the human gut symbiont Bacteroides thetaiotaomicron both in its native form and in complex with a mimic of the reaction intermediate. Mutagenesis and kinetics studies show that the bacterial enzyme, very similarly to its human counterpart, operates via an unusual 'substrate-assisted' catalytic mechanism, which will inform the rational design of enzyme inhibitors.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Torres, C.R. & Hart, G.W. Topography and polypeptide distribution of terminal N-acetylglucosamine residues on the surfaces of intact lymphocytes. Evidence for O-linked GlcNAc. J. Biol. Chem. 259, 3308–3317 (1984).
Kamemura, K., Hayes, B.K., Comer, F.I. & Hart, G.W. Dynamic interplay between O-glycosylation and O-phosphorylation of nucleocytoplasmic proteins: alternative glycosylation/phosphorylation of THR-58, a known mutational hot spot of c-Myc in lymphomas, is regulated by mitogens. J. Biol. Chem. 277, 19229–19235 (2002).
Cheng, X., Cole, R.N., Zaia, J. & Hart, G.W. Alternative O-glycosylation/O-phosphorylation of the murine estrogen receptor beta. Biochemistry 39, 11609–11620 (2000).
Lefebvre, T. et al. Effect of okadaic acid on O-linked N-acetylglucosamine levels in a neuroblastoma cell line. Biochim. Biophys. Acta 1472, 71–81 (1999).
Chou, C.F., Smith, A.J. & Omary, M.B. Characterization and dynamics of O-linked glycosylation of human cytokeratin 8 and 18. J. Biol. Chem. 267, 3901–3906 (1992).
Davis, L.I. & Blobel, G. Nuclear pore complex contains a family of glycoproteins that includes p62: glycosylation through a previously unidentified cellular pathway. Proc. Natl. Acad. Sci. USA 84, 7552–7556 (1987).
Dong, D.L., Xu, Z.S., Hart, G.W. & Cleveland, D.W. Cytoplasmic O-GlcNAc modification of the head domain and the KSP repeat motif of the neurofilament protein neurofilament-H. J. Biol. Chem. 271, 20845–20852 (1996).
Jackson, S.P. & Tjian, R. O-glycosylation of eukaryotic transcription factors: implications for mechanisms of transcriptional regulation. Cell 55, 125–133 (1988).
Zhang, F. et al. O-GlcNAc modification is an endogenous inhibitor of the proteasome. Cell 115, 715–725 (2003).
Lubas, W.A., Frank, D.W., Krause, M. & Hanover, J.A. O-Linked GlcNAc transferase is a conserved nucleocytoplasmic protein containing tetratricopeptide repeats. J. Biol. Chem. 272, 9316–9324 (1997).
Kreppel, L.K., Blomberg, M.A. & Hart, G.W. Dynamic glycosylation of nuclear and cytosolic proteins. Cloning and characterization of a unique O-GlcNAc transferase with multiple tetratricopeptide repeats. J. Biol. Chem. 272, 9308–9315 (1997).
Dong, D.L. & Hart, G.W. Purification and characterization of an O-GlcNAc selective N-acetyl-beta-D-glucosaminidase from rat spleen cytosol. J. Biol. Chem. 269, 19321–19330 (1994).
Gao, Y., Wells, L., Comer, F.I., Parker, G.J. & Hart, G.W. Dynamic O-glycosylation of nuclear and cytosolic proteins: cloning and characterization of a neutral, cytosolic beta-N-acetylglucosaminidase from human brain. J. Biol. Chem. 276, 9838–9845 (2001).
Shafi, R. et al. The O-GlcNAc transferase gene resides on the X chromosome and is essential for embryonic stem cell viability and mouse ontogeny. Proc. Natl. Acad. Sci. USA 97, 5735–5739 (2000).
Lehman, D.M. et al. A single nucleotide polymorphism in MGEA5 encoding O-GlcNAc-selective N-acetyl-beta-D glucosaminidase is associated with type 2 diabetes in Mexican Americans. Diabetes 54, 1214–1221 (2005).
Vosseller, K., Wells, L., Lane, M.D. & Hart, G.W. Elevated nucleocytoplasmic glycosylation by O-GlcNAc results in insulin resistance associated with defects in Akt activation in 3T3–L1 adipocytes. Proc. Natl. Acad. Sci. USA 99, 5313–5318 (2002).
Jinek, M. et al. The superhelical TPR-repeat domain of O-linked GlcNAc transferase exhibits structural similarities to importin alpha. Nat. Struct. Mol. Biol. 11, 1001–1007 (2004).
Zachara, N.E. et al. Dynamic O-GlcNAc modification of nucleocytoplasmic proteins in response to stress. A survival response of mammalian cells. J. Biol. Chem. 279, 30133–30142 (2004).
Toleman, C., Paterson, A.J., Whisenhunt, T.R. & Kudlow, J.E. Characterization of the HAT domain of a bifunctional protein with activatable O-GlcNAcase and HAT activities. J. Biol. Chem. 279, 53665–53673 (2004).
Henrissat, B. A classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem. J. 280, 309–316 (1991).
Xu, J. et al. A genomic view of the human-Bacteroides thetaiotaomicron symbiosis. Science 299, 2074–2076 (2003).
Knapp, S. et al. NAG-thiazoline, an N-acetyl-β-hexosaminidase inhibitor that implicates acetamido participation. J. Am. Chem. Soc. 118, 6804–6805 (1996).
Mohan, H. & Vasella, A. An improved synthesis of 2-acetamido-2-deoxy-D-gluconohydroximolactone (PUGNAc), a strong inhibitor of beta-N-acetylglucosaminidases. Helv. Chim. Acta 83, 114–118 (2000).
Mark, B.L. et al. Crystal structure of human beta-hexosaminidase B: understanding the molecular basis of Sandhoff and Tay-Sachs disease. J. Mol. Biol. 327, 1093–1109 (2003).
Mark, B.L. et al. Crystallographic evidence for substrate-assisted catalysis in a bacterial beta-hexosaminidase. J. Biol. Chem. 276, 10330–10337 (2001).
Holm, L. & Sander, C. Protein structure comparison by alignment of distance matrices. Trends Biochem. Sci. 20, 478–480 (1995).
Newstead, S.L., Watson, J.N., Bennet, A.J. & Taylor, G. Galactose recognition by the carbohydrate-binding module of a bacterial sialidase. Acta Crystallogr. D Biol. Crystallogr. 61, 1483–1491 (2005).
Sonnenburg, J.L. et al. Glycan foraging in vivo by an intestine-adapted bacterial symbiont. Science 307, 1955–1959 (2005).
Macauley, M.S., Whitworth, G.E., Debowski, A.W., Chin, D. & Vocadlo, D.J. O-GlcNAcase uses substrate-assisted catalysis: kinetic analysis and development of highly selective mechanism-inspired inhibitors. J. Biol. Chem. 280, 25313–25322 (2005).
Macauley, M.S., Stubbs, K.A. & Vocadlo, D.J. O-GlcNAcase catalyzes cleavage of thioglycosides without general acid catalysis. J. Am. Chem. Soc. 127, 17202–17203 (2005).
Vocadlo, D.J., Davies, G.J., Laine, R. & Withers, S.G. Catalysis by hen egg-white lysozyme proceeds via a covalent intermediate. Nature 412, 835–838 (2001).
Terwisscha van Scheltinga, A.C. et al. Stereochemistry of chitin hydrolysis by a plant chitinase/lysozyme and X-ray structure of a complex with allosamidin: evidence for substrate assisted catalysis. Biochemistry 34, 15619–15623 (1995).
Tews, I. et al. Bacterial chitobiase structure provides insight into catalytic mechanism and the basis of Tay-Sachs disease. Nat. Struct. Biol. 3, 638–648 (1996).
Markovic-Housley, Z. et al. Crystal structure of hyaluronidase, a major allergen of bee venom. Struct. Fold. Des. 8, 1025–1035 (2000).
Çetinbas, N., Macauley, M.S., Stubbs, K.A., Drapala, R. & Vocadlo, D.J. Identification of Asp174 and Asp175 as the key catalytic residues of human O-GlcNAcase by functional analysis of site-directed mutants. Biochemistry, 45, 3835–3844 (2006).
Stubbs, K.A., Zhang, N. & Vocadlo, D.J. A divergent synthesis of 2-acyl derivatives of PUGNAc yields selective inhibitors of O-GlcNAcase. Org. Biomol. Chem. 4, 839–845 (2006).
Roos, M.D. et al. Streptozotocin, an analog of N-acetylglucosamine, blocks the removal of O-GlcNAc from intracellular proteins. Proc. Assoc. Am. Physicians 110, 422–432 (1998).
Bolzan, A.D. & Bianchi, M.S. Genotoxicity of streptozotocin. Mutat. Res. 512, 121–134 (2002).
Rao, F.V. et al. Structural insights into the mechanism and inhibition of eukaryotic O-GlcNAc hydrolysis. EMBO J. (in the press).
Collaborative Computational Project, Number 4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50, 760–763 (1994).
Schneider, T.R. & Sheldrick, G.M. Substructure solution with SHELXD. Acta Crystallogr. D Biol. Crystallogr. 58, 1772–1779 (2002).
Murshudov, G.N., Vagin, A.A. & Dodson, E.J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 (1997).
Perrakis, A., Morris, R. & Lamzin, V.S. Automated protein model building combined with iterative structure refinement. Nat. Struct. Biol. 6, 458–463 (1999).
Emsley, P. & Cowtan, K. Coot. model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004).
Kraulis, P.J. MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr. 24, 946–950 (1991).
Esnouf, R.M. An extensively modified version of MolScript that includes greatly enhanced colouring capabilities. J. Mol. Graph. Model. 15, 132–134 (1997).
Wells, L. et al. Dynamic O-glycosylation of nuclear and cytosolic proteins: further characterization of the nucleocytoplasmic beta-N-acetylglucosaminidase, O-GlcNAcase. J. Biol. Chem. 277, 1755–1761 (2002).
Hansch, C. & Leo, A. Substituent Constants for Correlation Analysis in Chemistry and Biology (Wiley, New York, 1979).
Acknowledgements
R.J.D. thanks the Biotechnology and Biological Sciences Research Council (BBSRC) for a PhD fellowship. This work was supported by grants from the BBSRC to G.J.D. and from the Natural Sciences and Engineering Research Council of Canada (NSERC) and the Protein Engineering Network of Centres of Excellence to D.J.V. D.J.V. is supported as a Tier II Canada Research Chair in Chemical Glycobiology. M.S.M. thanks NSERC and the Michael Smith Foundation for Health Research for fellowships. We also thank A.J. Bennet for access to equipment. This work was funded by the BBSRC, the Royal Society of the UK and the NSERC.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Figure 1
Inhibitors used in the study of BtGH84 and key interactions within the enzyme active site (PDF 197 kb)
Supplementary Figure 2
Kinetic analyses of site-directed mutants of BtGH84 (PDF 124 kb)
Supplementary Figure 3
Electrostatic potential surface figure of the active center environment of BtGH84 (PDF 123 kb)
Rights and permissions
About this article
Cite this article
Dennis, R., Taylor, E., Macauley, M. et al. Structure and mechanism of a bacterial β-glucosaminidase having O-GlcNAcase activity. Nat Struct Mol Biol 13, 365–371 (2006). https://doi.org/10.1038/nsmb1079
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb1079
This article is cited by
-
Role of metagenomics in prospecting novel endoglucanases, accentuating functional metagenomics approach in second-generation biofuel production: a review
Biomass Conversion and Biorefinery (2023)
-
OGA is associated with deglycosylation of NONO and the KU complex during DNA damage repair
Cell Death & Disease (2021)
-
Enzymatic properties of β-N-acetylglucosaminidases
Applied Microbiology and Biotechnology (2018)
-
Structures of human O-GlcNAcase and its complexes reveal a new substrate recognition mode
Nature Structural & Molecular Biology (2017)
-
Genome-wide chemical mapping of O-GlcNAcylated proteins in Drosophila melanogaster
Nature Chemical Biology (2017)